FIG 7 .
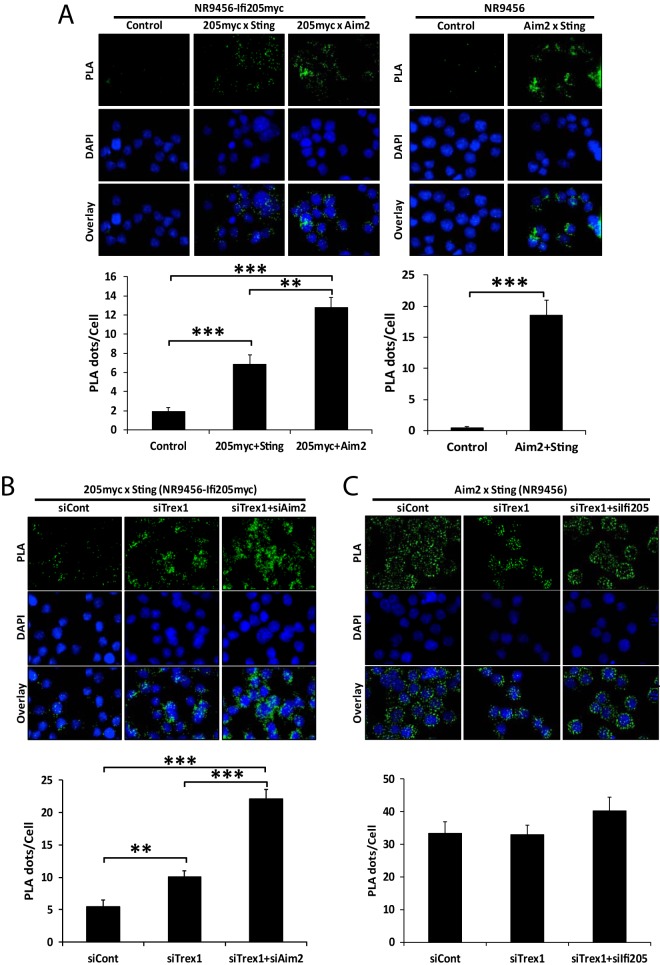
IFI205-STING interaction increases upon TREX1 and AIM2 depletion. (A) PLA for interactions of IFI205myc-AIM2, IFI205myc-STING, and AIM2-STING in NR9456-IFI205myc or NR9456 cells. The number of cells and images quantified for each condition in NR9456-IFI205myc cells are as follows: IFI205myc-Sting, 141 cells and 10 images; IFI205myc-Aim2, 101 cells and 6 images; control, 65 cells and 5 images. The number of cells and images quantified for each condition in NR9456 cells are as follows: control, 74 cells and 5 images; Aim2-Sting, 47 cells and 5 images. Controls for knockdowns are shown in Fig. S9A. (B) PLA for IFI205myc-STING interactions in NR9456-IFI205myc cells with knockdown of the genes indicated. Quantification: siCont, 102 cells and 5 images; siTrex1, 190 cells and 7 images; siTrex1+siAim2, 108 cells and 5 images. A representative image is shown. This experiment was repeated twice and gave similar results both times. (C) PLA for AIM2-STING interaction in NR9456 cells after knockdown of the genes indicated. Quantification: siCont, 84 cells and 4 images; siTrex1, 33 cells and 4 images; siTrex1+si205, 58 cells and 4 images. PLA dots were quantified and normalized to cell numbers based on DAPI staining with ImageJ software. The values shown are the mean ± the standard error of the mean of different pictures. **, P < 0.005; ***, P < 0.0005. (two-tailed t test). siCont, control siRNA.