Abstract
Aspergillus section Aspergillus (formerly the genus Eurotium) includes xerophilic species with uniseriate conidiophores, globose to subglobose vesicles, green conidia and yellow, thin walled eurotium-like ascomata with hyaline, lenticular ascospores. In the present study, a polyphasic approach using morphological characters, extrolites, physiological characters and phylogeny was applied to investigate the taxonomy of this section. Over 500 strains from various culture collections and new isolates obtained from indoor environments and a wide range of substrates all over the world were identified using calmodulin gene sequencing. Of these, 163 isolates were subjected to molecular phylogenetic analyses using sequences of ITS rDNA, partial β-tubulin (BenA), calmodulin (CaM) and RNA polymerase II second largest subunit (RPB2) genes. Colony characteristics were documented on eight cultivation media, growth parameters at three incubation temperatures were recorded and micromorphology was examined using light microscopy as well as scanning electron microscopy to illustrate and characterize each species. Many specific extrolites were extracted and identified from cultures, including echinulins, epiheveadrides, auroglaucins and anthraquinone bisanthrons, and to be consistent in strains of nearly all species. Other extrolites are species-specific, and thus valuable for identification. Several extrolites show antioxidant effects, which may be nutritionally beneficial in food and beverages. Important mycotoxins in the strict sense, such as sterigmatocystin, aflatoxins, ochratoxins, citrinin were not detected despite previous reports on their production in this section. Adopting a polyphasic approach, 31 species are recognized, including nine new species. ITS is highly conserved in this section and does not distinguish species. All species can be differentiated using CaM or RPB2 sequences. For BenA, Aspergillus brunneus and A. niveoglaucus share identical sequences. Ascospores and conidia morphology, growth rates at different temperatures are most useful characters for phenotypic species identification.
Key words: Ascomycota, Eurotiales, Aspergillaceae, Multi-gene phylogeny, Extrolites, Aspergillus proliferans, Eurotium amstelodami
Taxonomic novelties: Aspergillus aerius A.J. Chen, Frisvad & Samson; A. aurantiacoflavus Hubka, A.J. Chen, Jurjević & Samson; A. caperatus A.J. Chen, Frisvad & Samson; A. endophyticus Hubka, A.J. Chen, & Samson; A. levisporus Hubka, A.J. Chen, Jurjević & Samson; A. porosus A.J. Chen, Frisvad & Samson; A. tamarindosoli A.J. Chen, Frisvad & Samson; A. teporis A.J. Chen, Frisvad & Samson; A. zutongqii A.J. Chen, Frisvad & Samson
Introduction
Aspergillus subgenus Aspergillus, typified with A. glaucus (L.) Link, was introduced to include Aspergillus species with uniseriate conidiophore heads with hyaline, brownish or greenish stipes, and slightly inflated to subglobose vesicles and green conidia in mass (Gams et al. 1985). The subgenus contains two sections, namely sections Aspergillus (Aspergillus glaucus group Thom and Raper, 1941, Thom and Raper, 1945, Raper and Fennell, 1965) and Restricti (Aspergillus restrictus group Raper & Fennell 1965). The main difference of these two sections is that species in sect. Aspergillus readily produce a sexual state in culture (homothallic) and this sexual morph was, in the dual name nomenclature system, classified in the genus Eurotium (Malloch and Cain, 1972, Pitt, 1985). While the majority of species in sect. Restricti are asexually reproducing, one exception is A. halophilicus, which produces a eurotium-like sexual state (Christensen et al. 1959, Peterson et al. 2008). Peterson (2008) examined Aspergillus phylogenetically using β-tubulin (BenA), calmodulin (CaM), ID region of rDNA (ITS and partial LSU) and RNA polymerase II second largest subunit (RPB2), and showed that sections Aspergillus and Restricti formed a monophyletic subgenus Aspergillus. The monophyly of both sections was recently confirmed using a larger data set by Sklenář et al. (2017). Houbraken & Samson (2011) assessed relationships in the Trichocomaceae using a multigene phylogeny (RPB1, RPB2, Tsr1 and Cct8) and showed that Aspergillus and its sexual states formed a monophyletic clade closely related to Penicillium. This was again confirmed using a 25-gene phylogeny (Houbraken et al. 2014). Pitt & Taylor (2014) on the other hand, re-examined and analysed data from Houbraken and Samson (2011), and claimed that Penicillium could be included in a very broad concept of Aspergillus, which would only be monophyletic if Penicillium was included. This was partly caused by Aspergillus paradoxus, A. malodoratus and A. crystallinus, which were at that time still classified in Aspergillus, but currently in Penicillium. Similarly, Penicillium inflatum was combined in Aspergillus as Aspergillus inflatus (Visagie et al. 2014b, Samson et al. 2014). Furthermore, Pitt and Taylor, 2014, Pitt and Taylor, 2016 proposed to maintain the genus Eurotium for subgenus Aspergillus and subdivided Aspergillus into several smaller genera based on the corresponding sexual names. Most recently, Kocsubé et al. (2016) brought strong evidence that Aspergillus and Penicillium are monophyletic based on a robust multiple gene phylogenetic analyses and extrolite profiles. These findings rejected the hypothesis of Aspergillus being a paraphyletic genus (Pitt and Taylor, 2014, Pitt and Taylor, 2016), and were in agreement with the previous studies of Houbraken & Samson (2011) and Houbraken et al. (2014).
To avoid instability and nomenclatural confusion, the broad concept of Aspergillus was chosen by a majority of the International Commission of Penicillium and Aspergillus (ICPA) on April 11, 2012. Consequently, Hubka et al. (2013a) transferred all Eurotium taxa to Aspergillus. This treatment is widely accepted. Subsequently five species producing a eurotium-like sexual state, namely A. cumulatus, A. mallochii, A. megasporus, A. osmophilus and A. sloanii were introduced in sect. Aspergillus and Aspergillus names preferred over Eurotium by most authors (Asgari et al., 2014, Kim et al., 2014, Visagie et al., 2014a, Visagie et al., 2017). Before sequence data became widely available, the taxonomy of sect. Aspergillus was based on morphological characters. Ascospore pattern, shape and size were considered the most important characters distinguishing species, whereas the conidial apparatus and mycelial pigmentation provide valuable additional information (Thom and Raper, 1941, Thom and Raper, 1945, Raper and Fennell, 1965). Raper (1957) emphasized that in some strains the sexual state is dominating, while in others it is the asexual state, which has significant influence on the appearance of colonies. Blaser (1975) found that morphology and size of ascospores, surface ornamentation and colour of conidia are dependent on the temperature and water activity of cultivation media and thus reduced some species to synonyms. Samson (1979) compiled the sect. Aspergillus species published since Raper and Fennell's treatment in 1965, and synonymized six species under earlier names. Pitt (1985) reappraised the nomenclature and taxonomy of Eurotium (sect. Aspergillus), and accepted seven species based on the distinct nature of their ascospores. Kozakiewicz (1989) focused on scanning electron microscope (SEM) examinations of conidia and ascospores in her treatment of the group. Based on conidial ornamentations, four conidial morphotypes were identified, namely aculeate, tuberculate, lobate-reticulate and microtuberculate. Within each group, characters of equatorial crests, furrow and convex wall ornamentation are important diagnostic features. It was shown that some species previously considered conspecific according to light microscopy, e.g. A. cristatus (= Eurotium cristatum) and A. intermedius (= E. intermedium), show distinct conidial ornamentation in SEM and deserve to be recognized as separate species (Kozakiewicz 1989). Hubka et al. (2013a) studied the phylogeny of sect. Aspergillus based on ID region, BenA, CaM and RPB2 sequences, and accepted 17 species based on Genealogical Concordance Phylogenetic Species Recognition (GCPSR) approach.
Members of sect. Aspergillus are generally referred to as osmo-, xero- or halotolerant. They have a world-wide distribution and are common in indoor air, house dust, cereals, food products containing high concentrations of sugar, such as syrups, jams and jellies, salted meat products, semi-dry foods, feeds, leather goods and so on (Raper and Fennell, 1965, Blaser, 1975, Chelkowski et al., 1987, Pitt and Hocking, 2009, Samson et al., 2010, Greco et al., 2015). Species in this section are able to initiate growth at minimum moisture levels, thus establishing bridgeheads and facilitating the invasion of slightly less xerophilic molds (Semeniuk et al., 1947, Raper and Fennell, 1965, Kozakiewicz, 1989). Some species are involved in food manufacturing. Aspergillus cristatus or “Golden Flower Fungus” is used in the production of Fuzhuan brick tea in China (Wen, 1990, Qi and Sun, 1990, Xu et al., 2011); Aspergillus pseudoglaucus (= Eurotium repens) is used as a starter culture in the manufacturing of katsuobushi and fish sauce (Hayakawa et al., 1993, Dimici and Wada, 1994); A. pseudoglaucus, A. chevalieri and A. montevidensis are frequently isolated from meju (dried fermented soybeans); two newly described species A. cibarius and A. cumulatus are also isolated from meju or meju fermentation related environment (Hong et al., 2011, Hong et al., 2012, Kim et al., 2014). Aspergillus chevalieri, A. cristatus, A. glaucus, A. montevidensis, A. proliferans, A. pseudoglaucus and A. ruber have been reported from feedstuffs very often (Pitt and Hocking, 2009, Samson et al., 2010, Greco et al., 2015). These species have also been reported from other habitats and substrates. Aspergillus cristatus, A. glaucus, A. pseudoglaucus and A. ruber were listed as marine-derived (Li et al., 2004a, Li et al., 2004b, Li et al., 2006, Li et al., 2008a, Li et al., 2008b, Li et al., 2009, Li et al., 2010, Wang et al., 2006, Wang et al., 2007c, Du et al., 2007, Du et al., 2008, Du et al., 2012, Du et al., 2014, Smetanina et al., 2007, Tao et al., 2009, Gomes et al., 2012, Yan et al., 2012, Sun et al., 2013, Tang et al., 2014, Meng et al., 2015) and A. brunneus, A. chevalieri, A. cristatus, A. glaucus, A. intermedius, A. montevidensis, A. niveoglaucus, A. pseudoglaucus, A. ruber and A. xerophilus have been reported from soil (Guarro et al. 2012). However, sea-water and soil are matrices rather than habitats, usually of high water activity, where these fungi cannot grow or compete with other fungi. Species in sect. Aspergillus are not considered as important pathogens, although A. glaucus, A. chevalieri and A. montevidensis (= Eurotium amstelodami) have been reported from cases of superficial infections and sporadic invasive infections (de Hoog et al., 2000, Reboux et al., 2001, Roussel et al., 2004, Summerbell et al., 2005, Hubka et al., 2012).
Species of sect. Aspergillus produce many extrolites such as flavoglaucin, auroglaucin, isotetrahydroauroglaucin, neoechinulins A and B, echinulin, preechinulin, neochinulin E, epiheveadride and questin (Slack et al., 2009, Greco et al., 2015). Production of the potentially toxic echinulin has been reported from various strains of A. montevidensis (= E. amstelodami) (Allen, 1972, Gatti and Fuganti, 1979) and A. pseudoglaucus (= E. repens) (Smetanina et al. 2007). Other so-called toxins such as flavoglaucin and auroglaucin co-occur in various taxa of sect. Aspergillus, along with isotetrahydroauroglaucin in some A. montevidensis (= E. amstelodami) and A. ruber (= E. rubrum) strains (Slack et al. 2009). Interestingly, none of the compounds produced by these fungi have been classified as real mycotoxins, as the definition of the word mycotoxin is secondary metabolites (or extrolites) produced by filamentous fungi that are toxic to human beings and other vertebrates when introduced in small amounts via a natural route (orally, through pulmonary tract or skin) (Bennett & Klich 2003). On the other hand, the small molecule extrolites, such as dihydroauroglaucin (DAG), tetrahydroauroglaucin (TAG), anthraquinone derivatives, etc. produced by sect. Aspergillus species are antioxidant, and may even be beneficial to health (Ishikawa et al., 1985, Li et al., 2004a, Li et al., 2009, Miyake et al., 2009, Meng et al., 2016). Reports of sect. Aspergillus species producing true mycotoxins such as aflatoxins, ochratoxin A and sterigmatocystin were proved to be incorrect (Frisvad et al. 2007).
The aim of this study is to provide a taxonomic revision of sect. Aspergillus using a polyphasic approach. Phylogenetic relationships between sect. Aspergillus members were investigated using a combined data set (BenA, CaM and RPB2 sequences), and comparison of single-gene phylogenies was executed to determine tentative species boundaries based on genealogical concordance principle. Furthermore, phenotypic features including macro- and micro-morphology, ecophysiology and extrolite profiles are included in the polyphasic approach. Finally, the details on the identification of world-wide indoor environment strains have been included here.
Material and methods
Fungal strains
Strains used in this study were obtained from: 1) CBS, Culture Collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; 2) CGMCC, China General Microbiological Culture Collection Centre, Beijing, China; 3) NRRL, Agricultural Research Service Culture Collection, Peoria, Illinois, USA; 4) KACC, Korean Agricultural Culture Collection, Wanju, South Korea; 5) CCF, Culture Collection of Fungi, Prague, Czech Republic; 6) CCM (F-), Czech Collection of Microorganisms, Brno, Czech Republic; 7) IBT, the culture collection of Department of Biotechnology and Biomedicine, Technical University of Denmark; 8) DAOMC, Canadian Collection of Fungal Cultures, at the Ottawa Research and Development Centre – Agriculture and Agri-Food, Ottawa, Canada; and 9) BCCM/IHEM, Belgian Coordinated Collections of Microorganisms. Strains deposited in the working collection of the Applied and Industrial Mycology department (DTO) housed at the Westerdijk Fungal Biodiversity Institute, were also included in this study (Table 1).
Table 1.
Section Aspergillus strains used in phylogenetic analyses.
| Species | Strain nr.1 | Source | GenBank accession nr. |
|||
|---|---|---|---|---|---|---|
| ITS | BenA | CaM | RPB2 | |||
| Aspergillus aerius | CBS 141771T = DTO 241-G7 = IBT 34446 | The Netherlands, air treatment system in production plant, 2013, J. Houbraken | LT670916 | LT670990 | LT670991 | LT670992 |
| A. appendiculatus | CBS 374.75T = IMI 278374 = FRR 2793 = JCM 1566 = IBT 34507 | Switzerland, Stäfa, smoked sausage, 1971, P. Blaser | HE615132 | HE801333 | HE801318 | HE801307 |
| CBS 101746 = CGMCC 3.04673 (AS 3.4673) (ex-type of A. aridicola) | China, Tibet, sheep dung, H.Z. Kong & Z.T. Qi | HE615133 | HE801334 | HE801319 | HE801308 | |
| A. aurantiacoflavus | CBS 141930T = EMSL No. 2903 = CCF 5393 = DTO 355-I1 = IBT 34485 | USA, California, San Diego, baby carrier – backpack, 2015, Ž. Jurjević | LT670917 | LT670993 | LT670994 | LT670995 |
| EMSL No. 2693 = CCF 5391 = DTO 355-H7 | USA, IL, Chicago, rubber toy imported from China, 2015, Ž. Jurjević | LT670918 | LT670996 | LT670997 | LT670998 | |
| EMSL No. 3024 = CCF 5394 = DTO 355-H9 | USA, New Jersey, Cherry Hill, cake spread, 2015, Ž. Jurjević | LT670919 | LT670999 | LT671000 | LT671001 | |
| A. brunneus | CBS 112.26T = CBS 524.65 = IBT 5341 = NRRL 131 = NRRL 134 = ATCC 1021 = IFO 5862 = IMI 211378 = QM 7406 = Thom 4481 = Thom 5633.4 = WB 131 = CCF 5587 (neotype of A. echinulatus) | USA, California, fruit (Ficus carica), M.B. Church | EF652060 | EF651907 | EF651998 | EF651939 |
| DTO 357-A1 = KAS7575 | Canada, house dust, 2015, C.M. Visagie | LT670920 | LT671002 | LT671003 | LT671004 | |
| NRRL 133 = CCF 5586 | Unknown source, G. Smith | EF652061 | EF651908 | EF651999 | EF651940 | |
| NRRL 124 = CBS 113.27 = CCF 5585 (ex-type of A. medius) | Unknown source, W. McRae | EF652056 | EF651904 | EF651997 | EF651938 | |
| DTO 197-B3 = CBS 117328 | Canada, Manitoba, M. Desjardins | LT670921 | LT671005 | LT671006 | LT671007 | |
| A. caperatus | CBS 141774T = DTO 337-E6 = IBT 34451 | South Africa, Robben Island, soil, 2015, M. Meijer | LT670922 | LT671008 | LT671009 | LT671010 |
| A. chevalieri | CBS 522.65T = NRRL 78 = ATCC 16443 = IMI 211382 = NRRL A-7803 = Thom 4125.3 = WB 78 = IBT 5680 (neotype of A. equitis) | USA, coffee beans, 1916, C. Thom | EF652068 | EF651911 | EF652002 | EF651954 |
| NRRL 79 | USA, Indiana, Indianapolis, unknown source, Dr. Adams | EF652069 | EF651912 | EF652003 | EF651955 | |
| NRRL 4755 | USA, culture contamination, D.I. Fennell | EF652071 | EF651913 | EF652004 | EF651956 | |
| CCF 3291 = DTO 355-B6 | Czech Republic, Brno, rice, 1999, V. Ostrý | FR727116 | HE578085 | HE578099 | HE801314 | |
| CCF 1676 = DTO 355-B7 | Czech Republic, Prague, semolina, 1979, V. Muzikář | LT670923 | LT671011 | LT671012 | LT671013 | |
| CCF 4788 = KACC 47145 = DTO 355-B8 | South Korea, soybeans, 2012, D.H. Kim | LT670924 | LT671014 | LT671015 | LT671016 | |
| CGMCC 3.06132 = DTO 348-G5 | China, Tibet, soil, 2001 | LT670925 | LT671017 | LT671018 | LT671019 | |
| DTO 238-E3 | Unknown source, S. Suhendriani | LT670926 | LT671020 | LT671021 | LT671022 | |
| CBS 141769 = DTO 088-D7 | Madagascar, soil, 2008, J. Houbraken | LT670927 | LT671023 | LT671024 | LT671025 | |
| CGMCC 3.06492 = DTO 348-H3 | China, Yunnan, moldy peel, 2001 | LT670928 | LT671026 | LT671027 | LT671028 | |
| DTO 092-D3 | Madagascar, soil, 2008, J. Houbraken | LT670929 | LT671029 | LT671030 | LT671031 | |
| A. cibarius | KACC 46346T = DTO 197-D3 = IBT 32307 = CCF 4783 | South Korea, Icheon, meju, 2011, S.B. Hong | JQ918177 | JQ918180 | JQ918183 | JQ918186 |
| CCF 4098 = NRRL 62493 = DTO 354-I8 | Czech Republic, Prague, toenail of 56-year-old woman, 2010, P. Lysková | FR848828 | FR837968 | FR837973 | FR837979 | |
| CCF 4235 = NRRL 62492 = DTO 354-I7 | Czech Republic, Prague, toenail of 63-year-old man, 2012, P. Lysková | HE801341 | HE801330 | HE801324 | HE801313 | |
| CCF 4264 = DTO 354-I9 | Spain, Nerja cave, near Málaga, cave sediment (entrance chambre), 2011, A. Nováková | HE974462 | HE974436 | HE806186 | HE974428 | |
| KACC 49766 = CCF 4784 | The Netherlands, black bean, 2012, M. Meijer | LT670930 | LT671032 | LT671033 | LT671034 | |
| EMSL No. 1652 = CCF 5385 = DTO 355-G6 | USA, Pennsylvania, child's shoes, 2012, Ž. Jurjević | LT670931 | LT671035 | LT671036 | LT671037 | |
| EMSL No. 2498 = CCF 5383 = DTO 355-G7 | USA, Washington DC, chocolate glazed frosted donut, 2014, Ž. Jurjević | LT670932 | LT671040 | LT671041 | LT671042 | |
| EMSL No. 2865 = CCF 5384 = DTO 355-G8 | USA, California, Danville, chocolate chip cookies, 2015, Ž. Jurjević | LT670933 | LT671043 | LT671044 | LT671045 | |
| CGMCC 3.06498 = DTO 348-H7 | China, Hebei, soil, 2001 | LT670934 | LT671046 | LT671047 | LT671048 | |
| CGMCC 3.00450 = DTO 348-B5 | China, 1952 | LT670935 | LT671049 | LT671050 | LT671051 | |
| A. costiformis | CBS 101749T = CGMCC 3.04664 (AS 3.4664) = DTO 348-D8 = IBT 34456 = IBT 33662 | China, Hebei, moldy paper-box, 1992, H.Z. Kong | HE615136 | HE801338 | HE801320 | HE801309 |
| CCF 4097 = NRRL 62483 = DTO 354-I3 | Czech Republic, Prague, toenail of 5-year-old boy, 2010, P. Lysková | FR837960 | FR837970 | FR837974 | FR837978 | |
| DTO 326-B4 | The Netherlands, cellophane, 2015, J. Houbraken | LT670936 | LT671052 | LT671053 | LT671054 | |
| CGMCC 3.06520 = DTO 348-I5 | China, Hebei, moldy box, 2001 | LT670937 | LT671055 | LT671056 | LT671057 | |
| A. cristatus | CBS 123.53T = NRRL 4222 = ATCC 16468 = BCRC 33090 = FRR 1167 = IBT 5355 = IHEM 5619 = IMI 172280 = JCM 1569 = MUCL 15644 = NRRL 4222 = WB 4222 = CCF 5591 (ex-type of A. cristatellus) | South Africa, unknown, 1953, H.J. Swart | EF652078 | EF651914 | EF652001 | EF651957 |
| IHEM 2423 = DTO 355-B3 | Zaire, Kinshasa, soil, 1984 | LT670938 | LT671058 | LT671059 | LT671060 | |
| CCF 4701 = DTO 355-B1 | China, Hunan, tea block, 2013, Q.L. Pan & L. Wang | KF923732 | KF923737 | KF923741 | KF923734 | |
| CCF 4702 = DTO 355-B2 | China, Guangxi, tea block, 2013, Q.L. Pan & L. Wang | KF923733 | KF923739 | LT714711 | KF923736 | |
| CGMCC 3.06081 = DTO 348-E9 | China, Hubei, soil, 2001 | LT670939 | LT671061 | LT671062 | LT671063 | |
| A. cumulatus | KACC 47316T = DTO 303-D9 = IBT 34470 = IBT 33670 | South Korea, Anseong, rice straw used in meju fermentation | KF928303 | KF928297 | KF928300 | KF928294 |
| KACC 47513 = DTO 303-D8 | South Korea, air of a meju fermentation room | KF928304 | KF928298 | KF928301 | KF928295 | |
| KACC 47514 | South Korea, air of a meju fermentation room | KF928305 | KF928299 | KF928302 | KF928296 | |
| EMSL No. 2827 = CCF 5376 = DTO 355-G9 | USA, New York, Bronx, bedroom ceiling, 2015, Ž. Jurjević | LT670940 | LT671064 | LT671065 | LT671066 | |
| A. endophyticus | CBS 141766T = DTO 354-I2 = CCF 5345 = IBT 34511 | Czech Republic, Prague, Stromovka park, endophyte of Acer pseudoplatanus, 2013, I. Kelnarová | LT670941 | LT671067 | LT671068 | LT671069 |
| A. glaucus | CBS 516.65T = NRRL 116 = ATCC 16469 = DTO 197-A1 = IBT 32295 = IMI 211383 = LCP 64.1859 = Thom 5629.C = WB 116 | USA, Washington DC, unpainted board (K.B. Raper's residence), 1938, K.B. Raper | EF652052 | EF651887 | EF651989 | EF651934 |
| NRRL 117 = DTO 355-B4 = CCF 5582 (ex-type of A. mangini) | USA, Washington DC, unpainted board (K.B. Raper's basement), 1938, K.B. Raper | EF652053 | EF651888 | EF651990 | EF651935 | |
| EMSL No. 2529 = CCF 5381 = DTO 355-H1 | Puerto Rico, Bayamon, office, air, 2014, Ž. Jurjević | LT670942 | LT671070 | LT671071 | LT671072 | |
| NRRL 120 = 117.46 = CBS 532.65 = CCF 5583 (ex-type of A. umbrosus) | USA, coffee beans, 1925, F.A. McCormick | EF652054 | EF651889 | EF651991 | EF651936 | |
| NRRL 121 = DTO 355-B5 = CCF 5584 | Unknown source | EF652055 | EF651890 | EF651992 | EF651937 | |
| EMSL No. 3317 = CCF 5382 = DTO 355-H2 | USA, New York, Ulster Park, bedroom, settle plates, 2015, Ž. Jurjević | LT670943 | LT671073 | LT671074 | LT671075 | |
| A. intermedius | CBS 523.65T = NRRL 82 = ATCC 16444 = DSM 2830 = IBT 5677 = IMI 089278ii = IMI 89278 = LSHBBB 107 = LSHTM 107 = QM 7403 = Thom 5612.107 = WB 82 = CCF 5581 | UK, cotton yarn, 1927, G. Smith | EF652074 | EF651892 | EF652012 | EF651958 |
| NRRL 84 | Unknown source | EF652070 | EF651893 | EF652013 | EF651959 | |
| NRRL 4817 = DTO 355-B9 = IFO 5322 = IMI 313754 = JCM 23051 = CCF 5608 | Unknown country, butter | EF652072 | EF651894 | EF652014 | EF651960 | |
| NRRL 25823 | USA, IL, Peoria, soy protein, A.J. Moyer | EF652073 | EF651895 | EF652015 | EF651961 | |
| CBS 377.75 (ex-type of A. spiculosus) | Spain, Badajoz, soil, P. Blaser | HE974459 | HE974432 | HE974437 | HE974425 | |
| CCF 127 = DTO 354-I5 | China, industrial material, 1955, V. Zánová | HE578060 | HE974431 | HE578100 | HE974426 | |
| CCF 4681 = DTO 354-I6 | Czech Republic, Prague, sputum of 55-year-old woman, 2013, P. Lysková | LT670944 | LT671076 | LT671077 | LT671078 | |
| CCF 5377 = DTO 355-G5 | Czech Republic, Prague, air sampler, surgical operating room, 2014, A. Ešnerová | LT670945 | LT671079 | LT671080 | LT671081 | |
| CGMCC 3.03968 = DTO 348-D6 | China, unknown source, 1969 | LT670946 | LT671082 | LT671083 | LT671084 | |
| CGMCC 3.00664 = DTO 348-C1 | Czech Republic, unknown source, 1956 | LT670947 | LT671085 | LT671086 | LT671087 | |
| A. leucocarpus | CBS 353.68T = IBT 5350 = IMI 278375 = NRRL 3497 = QM 9365 = QM 9707 = CCF 5590 | Germany, Giessen, dried sausage, R. Hadlok | EF652087 | EF651925 | EF652023 | EF651972 |
| DTO 357-A2 = KAS7576 | Canada, house dust, 2015, C.M. Visagie | LT670948 | LT671088 | LT671089 | LT671090 | |
| DTO 174-I5 | Madagascar, vanilla sticks, 2012, J. Houbraken | LT670949 | LT671091 | LT671092 | LT671093 | |
| A. levisporus | CBS 141767T = DTO 355-G4 = EMSL No. 3211 = CCF 5378 = IBT 34512 | USA, Missouri, Saint Louis, bedroom, wood base, 2015, Ž. Jurjević | LT670950 | LT671094 | LT671095 | LT671096 |
| A. mallochii | CBS 141928T = DTO 357-A5 = KAS7618 = DAOMC 146054 | USA, California, San Mateo, pack rat dung, D. Malloch | KX450907 | KX450889 | KX450902 | KX450894 |
| CBS 141776 = DTO 343-G3 | The Netherlands, chocolat miroir, 2015 | KX450908 | KX450890 | KX450903 | KX450895 | |
| A. megasporus | CBS 141929T= DTO 356-H7 = KAS6176 = DAOMC 250799 | Canada, Nova Scotia, Wolfville, house dust, 2015, C.M. Visagie | KX450910 | KX450892 | KX450905 | KX450897 |
| CBS 141772 = DTO 048-I3 | The Netherlands, Dutch chocolate butter, 2007, M. Meijer | KX450911 | KX450893 | KX450906 | KX450898 | |
| DTO 356-H1 = KAS5973 = DAOMC 250800 | Canada, New Brunswick, Little Lepreau, house dust, 2015, C.M. Visagie | KX450909 | KX450891 | KX450904 | KX450896 | |
| A. montevidensis | CBS 491.65T = NRRL 108 = ATCC 10077 = IBT 5685 = IHEM 3337 = IMI 172290 = NRRL 109 = QM 7423 = Thom 5290 = Thom 5633.24 = WB 108 | Uruguay, Montevideo, tympanic membrane of human ear, 1932, R.V. Talice & J.E. MacKinnon | EF652077 | EF651898 | EF652020 | EF651964 |
| NRRL 89 | Unknown source | EF652075 | EF651896 | EF652016 | EF651962 | |
| NRRL 90 = CBS 518.65 (ex-type of A. hollandicus) | USA, unknown source, ∼1910 | EF652076 | EF651897 | EF652017 | EF651963 | |
| NRRL 4716 | USA, Missouri, Columbia, candied grapefruit rind, D.I. Fennell | EF652079 | EF651899 | EF652018 | EF651965 | |
| NRRL 25850 | USA, IL, Peoria, refrigerated bread dough, R. Graves | EF652082 | EF651900 | EF652021 | EF651966 | |
| NRRL 35697 | USA, IL, Chicago, nasal swab | EF652084 | EF651902 | EF652022 | EF651968 | |
| NRRL A-13891 = CBS 410.65 (ex-type of A. heterocaryoticus) | Mexico, Oryza sativa kernel, 1963, C.R. Benjamin | EU021619 | EU021670 | EU021687 | EU021659 | |
| CBS 651.74 = ATCC 24717 = IMI 174724 = VKM F-1760 (ex-type of A. vitis) | Kazakhstan, Alma-Ata, ex grapes, 1968, L.A. Beljakova | HE974460 | HE974433 | HE974441 | HE974424 | |
| CCF 3998 | Czech Republic, Prague, neck skin of 78-year-old woman, 2008, M. Skořepová | FR727117 | HE974434 | FR751447 | HE974418 | |
| CCF 4069 | Czech Republic, heel skin of 32-year-old man, Prague, 2007, M. Skořepová | FR839679 | FR775356 | HE974440 | HE974419 | |
| CCF 4070 | Czech Republic, fingernail of 32-year-old woman, Prague, 2007, M. Skořepová | FR848825 | FR775335 | FR751442 | HE974420 | |
| CCF 4071 | Czech Republic, Prague, thigh and neck skin of 42-year-old woman, 2010, P. Lysková | FR839680 | HE974435 | FR751449 | HE974421 | |
| CCF 4248 | Czech Republic, Skrbeň, window sill, 1997, A. Kubátová | HE974461 | HE801339 | HE974442 | HE974422 | |
| EMSL No. 2934 = CCF 5379 = DTO 355-H3 | USA, PA, Mahanoy City, bedroom, settle plates, 2015, Ž. Jurjević | LT670951 | LT671097 | LT671098 | LT671099 | |
| CBS 111.52 = DTO 351-C9 | Suriname, plywood, M.B. Schol-Schwarz | LT670952 | LT671100 | LT671101 | LT671102 | |
| DTO 147-I4 | Hungary, indoor air, 2014, M. Meijer | LT670953 | LT671103 | LT671104 | LT671105 | |
| CGMCC 3.03888 = DTO 348-D3 | China, mite, 1969 | LT670954 | LT671106 | LT671107 | LT671108 | |
| A. neocarnoyi | CBS 471.65T = NRRL 126 = ATCC 16924 = IBT 6016 = IMI 172279 = LSHTM A32 = QM 7402 = Thom 5612.A32 = WB 126 = DTO 196-H6 = CCF 5588 | Unknown source, P. Biourge | EF652057 | EF651903 | EF651985 | EF651942 |
| EXF-10029 = DTO 357-E2 | Slovenia, Ljubljana, Slovene Ethnographic museum, air at the sampling of shaman statue originating from Mali, 2016, P. Zalar | LT670955 | LT671109 | LT671110 | LT671111 | |
| A. niveoglaucus | CBS 114.27T = CBS 517.65 = NRRL 127 = ATCC 10075 = BCRC 33096 = CGMCC 3.04374 = FRR 927 = IBT 5356 = IMI 32050 = JCM 1578 = LSHBA 16 = NRRL 129 = NRRL 130 = QM 1977 = Thom 5612.A16 = Thom 5633 = Thom 5633.7 = Thom 7053.2 = UAMH 6591 = WB 127 = WB 130 = CCF 5589 (lectotype of A. glauconiveus) | Unknown source, A. Blochwitz | EF652058 | EF651905 | EF651993 | EF651943 |
| NRRL 128 | Unknown source, G. Smith | EF652059 | EF651906 | EF651994 | EF651944 | |
| NRRL 136 | Unknown source, G. Smith | EF652062 | EF651909 | EF651995 | EF651945 | |
| NRRL 137 | Unknown source | EF652063 | EF651910 | EF651996 | EF651946 | |
| CCF 4191 = DTO 355-C1 | Spain, Andalusia, Málaga, Cueva del Tesoro, cave sediment from the cave wall, 2010, A. Nováková | HE801344 | HE801332 | HE974438 | HE974427 | |
| CCM F-530 = CCF 4038 | Czech Republic, garlic, L. Marvanová | HE578069 | HE578086 | HE578092 | HE578114 | |
| EMSL No. 2211 = CCF 5380 = DTO 355-H8 | USA, Montana, Great Falls, air of bathroom, 2013, Ž. Jurjević | LT670956 | LT671112 | LT671113 | LT671114 | |
| IHEM 1811 = DTO 355-C3 | Belgium, Namur, indoor air, 1983 | LT670957 | LT671115 | LT671116 | LT671117 | |
| CBS 101750 = CGMCC 3.04665 (AS 3.4665) = DTO 197-B4 (ex-type of A. parviverruculosus) | China, Hebei, soil | HE615135 | HE801331 | HE801323 | HE801312 | |
| CCF 4787 = KACC 47144 = DTO 355-C4 | South Korea, soybeans, 2012, D.H. Kim | LT670958 | LT671118 | LT671119 | LT671120 | |
| CCF 4790 = KACC 47147 = DTO 355-C5 | South Korea, soybeans, 2012, D.H. Kim | LT670959 | LT671121 | LT671122 | LT671123 | |
| CGMCC 3.06092 = DTO 348-F3 | China, Guangdong, cashew Kernel, 2001 | LT670960 | LT671124 | LT671125 | LT671126 | |
| A. osmophilus | CBS 134258T = IRAN 2090C = DTO 354-C1 | Iran, East Azerbaijan province, Marand, Triticum aestivum leaf, 2006, B. Asgari | KC473921 | LT671127 | LT671128 | LT671129 |
| A. porosus | CBS 141770T = DTO 262-D7 = IBT 34443 | Turkey, soil, 2013, A. Yoltas | LT670961 | LT671130 | LT671131 | LT671132 |
| DTO 308-D1 | Turkey, soil, 2014, R. Demirel | LT670962 | LT671133 | LT671134 | LT671135 | |
| CBS 375.75 = DTO 197-C4 | Israel, Arachis hypogaea fruit, P. Blaser | LT670963 | LT671136 | LT671137 | LT671138 | |
| DTO 262-D4 | Turkey, soil, 2013, A. Yoltas | LT670964 | LT671139 | LT671140 | LT671141 | |
| DTO 262-D2 | Turkey, soil, 2013, A. Yoltas | LT670965 | LT671142 | LT671143 | LT671144 | |
| A. proliferans | CBS 121.45T = NRRL 1908 = IBT 6213 = IMI 016105ii = IMI 016105iii = IMI 16105 = LSHB BB.82 = MUCL 15625 = NCTC 6546 = QM 7462 = UC 4303 = WB 1908 = CCF 5580 | UK, Manchester, cotton yarn, G. Smith | EF652064 | EF651891 | EF651988 | EF651941 |
| DTO 322-A2 | The Netherlands, egg waffles, 2014, M. Meijer | LT670966 | LT671145 | LT671146 | LT671147 | |
| CCF 4192 = DTO 355-C6 | Spain, Andalusia, Aracena, Gruta de la Maravillas, cave sediment, 2010, A. Nováková | HE615128 | HE801328 | HE801316 | HE801305 | |
| NRRL 114 = DTO 355-C7 = CCF 5579 | USA, Massachusetts, unknown source | EF652051 | EF651886 | EF651987 | EF651933 | |
| CCF 4096 = NRRL 62482 = DTO 355-C8 | Czech Republic, Prague, palm skin, 28-year-old woman, 2008, M. Skořepová | FR848827 | FR775375 | HE650908 | HE801303 | |
| CCF 4115 = NRRL 62497 = DTO 355-C9 | Czech Republic, Prague, toenail of 64-year-old man, 2010, P. Lysková | FR851850 | FR851855 | HE578090 | HE578107 | |
| CCF 4146 = NRRL 62494 = DTO 355-D1 | Czech Republic, Prague, toenail of 48-year-old man, 2011, P. Lysková | HE578067 | HE578076 | HE650909 | HE801304 | |
| NRRL 71 = DTO 355-D2 = CCF 5578 | USA, Maryland, leafhoppers, V.K. Charles | EF652047 | EF651885 | EF651986 | EF651932 | |
| CCF 4232 | Czech Republic, Opava, stuffed bird, 2010, M. Polásek | HE615129 | HE801329 | HE801317 | HE801306 | |
| EMSL No. 2207 = CCF 5395 = DTO 355-H5 | USA, Pennsylvania, Yardley, air of living room, 2013, Ž. Jurjević | LT670967 | LT671148 | LT671149 | LT671150 | |
| EMSL No. 2791 = CCF 5392 = DTO 355-H6 | USA, New York, Troy, basement, settle plates, 2015, Ž. Jurjević | LT670968 | LT671151 | LT671152 | LT671153 | |
| CCF 4789 = KACC 47146 = DTO 355-D3 | South Korea, soybeans, 2012, D.H. Kim | LT670969 | LT671154 | LT671155 | LT671156 | |
| A. pseudoglaucus | CBS 123.28T = NRRL 40 = ATCC 10066 = IBT 5353 = IMI 016122 = IMI 016122ii = LSHBA 19 = MUCL 15624 = QM 7463 = Thom 5343 = WB 40 (lectotype of A. glaucoaffinis) | Unknown source, 1929, A. Blochwitz | EF652050 | EF651917 | EF652007 | EF651952 |
| NRRL 13 = CBS 529.65 (ex-type of A. reptans) | France, Prunus domestica, da Fonseca | EF652048 | EF651915 | EF652005 | EF651950 | |
| NRRL 17 | USA, wrist skin | EF652049 | EF651916 | EF652006 | EF651951 | |
| NRRL 25865 | Japan, Tokyo, unknown source, T. Ohtsuki | EF652065 | EF651918 | EF652008 | EF651953 | |
| CBS 101747 = CGMCC 3.04674 (AS 3.4674) (ex-type of A. fimicola) | China, Tibet, animal dung | HE615130 | HE801335 | HE801321 | HE801310 | |
| CBS 379.75 (ex-type of A. glaber) | Switzerland, Zuoz, Vaccinium myrtillus leaf, P. Blaser | HE615131 | HE801336 | HE801322 | HE801311 | |
| CCF 3283 | Czech Republic, Prague, 2002, A. Kubátová | FR727114 | FR775360 | HE974439 | HE578110 | |
| CCF 4011 | Czech Republic, Prague, back skin of 39-year-old woman, 2008, M. Skořepová | FR839678 | FR775358 | FR751446 | HE578111 | |
| EMSL No. 1780 = CCF 5388 = DTO 355-I2 | USA, Pennsylvania, floor swab, 2012, Ž. Jurjević | LT670970 | LT671157 | LT671158 | LT671159 | |
| EMSL No. 2779 = CCF 5389 = DTO 355-I3 | USA, Florida, Melbourne, vent, settle plates, 2015, Ž. Jurjević | LT670971 | LT671160 | LT671161 | LT671162 | |
| EMSL No. 2809 = CCF 5386 | USA, New York, Endicott, office, settle plates, 2015, Ž. Jurjević | LT670972 | LT671163 | LT671164 | LT671165 | |
| EMSL No. 2474 = CCF 5387 = DTO 355-I4 | USA, New Jersey, Piscataway, air, basement, 2014, Ž. Jurjević | LT670973 | LT671166 | LT671167 | LT671168 | |
| EMSL No. 2853 = CCF 5390 = DTO 355-I5 | USA, Missouri, St. Louis, cheddar cheese, 2015, Ž. Jurjević | LT670974 | LT671169 | LT671170 | LT671171 | |
| CBS 108961 = DTO 351-D2 | The Netherlands, Woerden, parmezan cheese, J. Houbraken | LT670975 | LT671172 | LT671173 | LT671174 | |
| DTO 147-G3 | Hungary, indoor air, 2010 | LT670976 | LT671175 | LT671176 | LT671177 | |
| CGMCC 3.00460 = DTO 348-B9 | China, tea, 1952 | LT670977 | LT671178 | LT671179 | LT671180 | |
| A. ruber | CBS 530.65T = NRRL 52 = ATCC 16441 = IBT 5453 = IMI 211380 = JCM 22942 = QM 1973 = Thom 5599B = WB 52 | Unknown source | EF652066 | EF651920 | EF652009 | EF651947 |
| NRRL 76 | Unknown source, G. Smith | EF652067 | EF651921 | EF652011 | EF651948 | |
| NRRL 5000 = CBS 464.65 (ex-type of A. athecius) | UK, coffee beans, 1965, E. Yuill | EF652080 | EF651922 | EF652010 | EF651949 | |
| CBS 101748 = CGMCC 3.04632 (AS 3.4632) (ex-type of A. tuberculatus) | China, Shanxi, soil | HE615134 | HE801337 | HE801325 | HE801315 | |
| CCF 2920 | Czech Republic, Nymburk, malt dust, 1993, A. Kubátová | FR727112 | FR775357 | FR751444 | HE974430 | |
| CCF 4377 | Czech Republic, Prague, toenail of 60-year-old woman, 2011, P. Lysková | HE578065 | HE578087 | HE578098 | LT671190 | |
| CBS 104.18 = DTO 351-C4 | Unknown source, 1918, O. Goethals | LT670978 | LT671181 | LT671182 | LT671183 | |
| DTO 238-C4 | Unknown source, Rahmawati | LT670979 | LT671184 | LT671185 | LT671186 | |
| CGMCC 3.00457 = DTO 348-B6 | China, tea, 1952 | LT670980 | LT671187 | LT671188 | LT671189 | |
| A. sloanii | CBS 138177T = DTO 245-A1 = IBT 34509 = CCF 4927 | UK, Middlesex, house dust, 2010, E. Whitfield & K. Mwange | KJ775540 | KJ775074 | LT671038 | KX463365 |
| CBS 138176 = DTO 244-I8 = CCF 4926 | UK, Middlesex, house dust, 2010, E. Whitfield & K. Mwange | KJ775539 | KJ775073 | LT671039 | KX463364 | |
| CBS 138231 = DTO 245-A6 | UK, Middlesex, house dust, 2010, E. Whitfield & K. Mwange | KJ775541 | KJ775075 | KJ775311 | KX450899 | |
| CBS 138178 = DTO 245-A8 | UK, Middlesex, house dust, 2010, E. Whitfield & K. Mwange | KJ775542 | KJ775076 | KJ775313 | KX450900 | |
| CBS 138179 = DTO 245-A9 | UK, Middlesex, house dust, 2010, E. Whitfield & K. Mwange | KJ775543 | KJ775077 | KJ775314 | KX450901 | |
| A. tamarindosoli | CBS 141775T = DTO 054-A8 = IBT 34432 | Thailand, Hua Hin, soil under tamarind, 2007, R.A. Samson & J. Houbraken | LT670981 | LT671191 | LT671192 | LT671193 |
| A. teporis | CBS 141768T = DTO 058-E5 = IBT 34513 | The Netherlands, heat treated corn kernels, 2008, M. Meijer | LT670982 | LT671194 | LT671195 | LT671196 |
| A. tonophilus | CBS 405.65T = NRRL 5124 = ATCC 16440 = ATCC 36504 = IBT 21230 = IMI 108299 = QM 8599 = WB 5124 = CCF 5592 | Japan, Tokyo, binocular lens, T. Ohtsuki | EF652081 | EF651919 | EF652000 | EF651969 |
| DTO 356-H6 = KAS6175 | Canada, house dust, 2015, C.M. Visagie | LT670915 | LT671197 | LT671198 | LT671199 | |
| CCF 4785 = KACC 45365 = DTO 355-A2 | South Korea, meju, 2012, S.B. Hong | LT670984 | LT671200 | LT671201 | LT671202 | |
| CCF 4786 = KACC 47150 = DTO 355-A1 | South Korea, soybeans, 2012, D.H. Kim | LT670985 | LT671203 | LT671204 | LT671205 | |
| A. xerophilus | CBS 938.73T = NRRL 6131 = IBT 5429 = IBT 5489 = IBT 34503 = DTO 083-A2 = CCF 5593 | Egypt, Western desert, desert soil, J. Mouchacca | EF652085 | EF651923 | EF651983 | EF651970 |
| NRRL 6132 = CBS 755.74 | Egypt, Western desert, desert soil, J. Mouchacca | EF652086 | EF651924 | EF651984 | EF651971 | |
| A. zutongqii | CBS 141773T = CGMCC 3.13917 = DTO 349-E1 = IBT 34450 | China, Beijing, peanut shell, 2008, L. Wang | LT670986 | LT671206 | LT671207 | LT671208 |
| CGMCC 3.06103 = DTO 348-F7 | China, Ningxia, 2001 | LT670987 | LT671209 | LT671210 | LT671211 | |
| CGMCC 3.03980 = DTO 348-D7 | China, 1969, Z.T. Qi | LT670988 | LT671212 | LT671213 | LT671214 | |
| CGMCC 3.03961 = DTO 348-D5 | China, ocular lens, 1969, Z.T. Qi | LT670989 | LT671215 | LT671216 | LT671217 | |
Culture collection designations: CBS, Culture Collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; CGMCC, China General Microbiological Culture Collection Centre, Beijing, China; NRRL, Agricultural Research Service Culture Collection, Peoria, Illinois, USA; KACC, Korean Agricultural Culture Collection, Wanju, South Korea; CCF, Culture Collection of Fungi, Prague, Czech Republic; CCM (F-), Czech Collection of Microorganisms, Brno, Czech Republic; IBT, culture collection of the DTU Systems Biology, Lyngby, Denmark; DAOMC, Canadian Collection of Fungal Cultures, at the Ottawa Research and Development Centre – Agriculture and Agri-Food, Ottawa, Canada; BCCM/IHEM, Belgian Coordinated Collections of Microorganisms; DTO, working collection of the Applied and Industrial Mycology department (DTO) housed at the Westerdijk Fungal Biodiversity Institute.
For newly isolated strains from the indoor environments, different isolation techniques were used. House dust samples were collected as described in Amend et al. (2010) and isolated using a modified dilution-to-extinction method (Visagie et al. 2014a). Air samples were collected approximately 1 m above the ground with a viable impaction sampler (MAS 100 Merck) (Peterson & Jurjević 2013). Indoor surfaces (walls, ceilings) were sampled with the swab (Greiner Bio-One, Alphen aan de Rijn, the Netherlands). For air and swab sampling, standard microbiological techniques were used for isolation. Malt extract agar (MEA) with chloramphenicol and Dichloran 18 % glycerol agar (DG18) were used as isolation media.
DNA extraction, PCR amplification and sequencing
Strains were grown for 1 wk on M40Y prior to DNA extraction. DNA was extracted using the Ultraclean™ Microbial DNA isolation Kit (MoBio, Solana Beach, U.S.A.) or the ArchivePure DNA yeast and Gram2+ kit (5 PRIME Inc., Gaithersburg, MD) according to manufacturer instructions updated by Hubka et al. (2015). Target loci, i.e. ITS, BenA, CaM and RPB2, were amplified using primer combination listed in Table 2. PCR product purification followed the protocol described by Réblová et al. (2016). Automated sequencing was performed at the Macrogen Sequencing Service (Amsterdam, the Netherlands) using same primers used in PCR.
Table 2.
Primers used in this study for amplification and sequencing.
| Locus | Primer | Amplification | Annealing temp (°C) | Cycles | Orientation | Sequence (from 5′ to 3′) | References |
|---|---|---|---|---|---|---|---|
| ITS | V9G (General, Gen.) | Standard | 55 (alt. 52) | 35 | Forward | TTACGTCCCTGCCCTTTGTA | de Hoog & Gerrits van den Ende (1998) |
| LS266 (Gen.) | Reverse | GCATTCCCAAACAACTCGACTC | Masclaux et al. (1995) | ||||
| ITS1 (Alternative, Alt.) | Forward | TCCGTAGGTGAACCTGCGG | White et al. (1990) | ||||
| ITS4 (Alt.) | Reverse | TCCTCCGCTTATTGATATGC | White et al. (1990) | ||||
| BenA | Bt2a (Gen.) | Standard | 55 (alt. 52) | 35 | Forward | GGTAACCAAATCGGTGCTGCTTTC | Glass & Donaldson (1995) |
| Bt2b (Gen.) | Reverse | ACCCTCAGTGTAGTGACCCTTGGC | Glass & Donaldson (1995) | ||||
| T10 (Alt.) | Forward | ACGATAGGTTCACCTCCAGAC | O'Donnell & Cigelnik (1997) | ||||
| Ben2F (Alt.) | Forward | TCCAGACTGGTCAGTGTGTAA | Hubka & Kolařík (2012) | ||||
| CaM | CMD5 (Gen.) | Standard | 55 (alt. 52) | 35 | Forward | CCGAGTACAAGGAGGCCTTC | Hong et al. (2005) |
| CMD6 (Gen.) | Reverse | TTTYTGCATCATRAGYTGGAC | Hong et al. (2005) | ||||
| CF1L (Alt.) | Forward | GCCGACTCTTTGACYGARGAR | Peterson (2008) | ||||
| CF1M (Alt.) | Forward | AGGCCGAYTCTYTGACYGA | Peterson (2008) | ||||
| CF4 (Alt.) | Reverse | TTTYTGCATCATRAGYTGGAC | Peterson (2008) | ||||
| RPB2 | fRPB2-5F (Gen.) | Standard | 55 (alt. 52 or 50) | 35 | Forward | GAYGAYMGWGATCAYTTYGG | Liu et al. (1999) |
| fRPB2-7CR (Gen.) | Reverse | CCCATRGCTTGYTTRCCCAT | Liu et al. (1999) | ||||
| fRPB2ResF100 (Alt.) | Touch-up | 44-46-48 | 5-5-30 | Forward | TGAARTAYGCICTTGCYAC | Sklenář et al. (2017) | |
| fRPB2ResR950 (Alt.) | Reverse | CARTGYGTCCADGTRTGKGC | Sklenář et al. (2017) | ||||
| RPB2-F50-CanAre (Alt.) | Touch-down | 65-64-63-62-61-60-55 | 1-1-1-1-1-1-38 | Forward | TTGAACATTGGTGTCAAGGC | Jurjević et al. (2015) |
Phylogenetic analysis
Sequences were inspected and assembled in BioEdit v.7.1.8 (http://www.mbio.ncsu.edu/bioedit/bioedit.html). Sequence alignments were performed using the FFT-NSi strategy implemented in MAFFT v.7 (Katoh & Standley 2013). Alignment characteristics are listed in Table 3. Maximum likelihood (ML) trees were constructed with IQ-TREE v. 1.4.0 (Nguyen et al. 2015). Optimal partitioning scheme and substitution models were selected using PartitionFinder v1.1.0 (Lanfear et al. 2012) with setting allowing introns, exons and codon positions to be independent datasets. The Bayesian information criterion was used to determine the model that best fits the data. Proposed partitioning schemes and substitution models for each dataset are listed in Table 4. Support values at branches were obtained from 1 000 bootstrap replicates. The trees were rooted with Hamigera avellanea NRRL 1938. MrBayes 3.2.2 (Ronquist et al. 2012) was used to calculate Bayesian posterior probabilities (PP). Optimal partitioning scheme and substitution models were selected using PartitionFinder v1.1.0 as described above. The analyses ran for 107 generations, two parallel runs with four chains each were used, every 1 000th tree was retained, and the first 25 % of trees were discarded as burn-in. All alignments are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.7hn1j.
Table 3.
Overview of alignments characteristics used for phylogenetic analyses (excluding outgroup).
| ITS | BenA | CaM | RPB2 | BenA + CaM + RPB2 | |
|---|---|---|---|---|---|
| Length (bp) | 538 | 402 | 710 | 969 | 2 081 |
| Variable position | 76 | 164 | 284 | 286 | 734 |
| Parsimony informative sites | 52 | 149 | 251 | 243 | 643 |
Table 4.
Partition-merging results and best substitution model for each partition according to Bayesian information criterion (BIC) as proposed by PartitionFinder v1.1.0.
| Dataset | Phylogenetic method | Partitioning scheme (substitution model) |
|---|---|---|
| ITS | ML | ITS1 + ITS2 (HKY+G); 5.8S (JC+I) |
| BI | ITS1 + ITS2 (HKY+G); 5.8S (JC+I) | |
| BenA | ML | introns (K80+G); 1st codon positions (JC+I); 2nd codon positions (JC); 3rd codon positions (K81uf+G) |
| BI | introns (K80+G); 1st codon positions (JC+I); 2nd codon positions (JC); 3rd codon positions (HKY+G) | |
| CaM | ML | introns (HKY+I+G); 1st codon positions (TrN+I); 2nd codon positions (F81); 3rd codon positions (TrN+G) |
| BI | introns (HKY+I+G); 1st codon positions (HKY+I); 2nd codon positions (F81); 3rd codon positions (GTR+G) | |
| RPB2 | ML | 1st codon positions (TrN+I+G); 2nd codon positions (JC+I); 3rd codon positions (TrNef+G) |
| BI | 1st + 2nd codon positions (K80+I+G); 3rd codon positions (HKY+G) | |
| BenA + CaM + RPB2 | ML | BenA + CaM introns (K81uf+I+G); 1st codon positions of BenA + CaM + RPB2 (TrN+I+G); 2nd codon positions of BenA + CaM + RPB2 (F81+I); 3rd codon positions of BenA + CaM (GTR+G); 3rd codon positions of RPB2 (TrNef+G) |
| BI | BenA + CaM introns (HKY+I+G); 1st codon positions of BenA + CaM + RPB2 (GTR+I+G); 2nd codon positions of BenA + CaM + RPB2 (F81+I); 3rd codon positions of BenA + CaM (GTR+G); 3rd codon positions of RPB2 (HKY+G) |
Morphological analysis
Macroscopic characters were studied on agar media Czapek yeast autolysate agar (CYA), CYA with 20 % sucrose agar (CY20S), CYA supplemented with 5 % NaCl (CYAS), Malt extract agar (MEA; Oxoid), MEA with 40 % sucrose agar (M40Y), MEA with 60 % sucrose agar (M60Y), MEA supplemented with 10 % NaCl (MEA10S) and Dichloran 18 % glycerol agar (DG18). Trace elements (0.1 g ZnSO4·7H2O and 0.5 g CuSO4·5H2O in 100 ml distilled water) were added to all media to obtain stable pigment production and consistent conidial colours (Smith, 1949, Samson et al., 2014). Isolates were inoculated at three points on 90 mm plates and incubated for 7 d at 25 °C in darkness. In addition, CY20S and M60Y plates were incubated at 30 °C and 37 °C, respectively. After 7 d of incubation, colony diameters were recorded. Colony texture, degree of sporulation, obverse and reverse colony colours, production of soluble pigments, exudates and ascomata were determined. Colour codes used in description refer to Rayner (1970).
Light microscope preparations were made from 1 wk old colonies grown on M40Y. Ascomata, asci and ascospores were observed after 2 or 4 wks. Lactic acid (60 %) was used as mounting fluid. Ethanol (96 %) was used to remove excess conidia and prevent air bubbles. A Zeiss Stereo Discovery V20 dissecting microscope and Zeiss AX10 Imager A2 light microscope both equipped with Nikon DS-Ri2 cameras and NIS-Elements D v 4.50 software were used to capture digital images.
Cryo Scanning Electron Microscopy (cryo-SEM) observations of ascospores were prepared based on Chen et al. (2016), alternatively, an osmium tetroxide method was used for fixation as described by Hubka et al. (2013b). To prevent conidia collapsing, agar blocks containing conidial structures were snap-frozen and observed as described in Visagie et al. (2013).
Extrolite analysis
Strains were incubated on DG18, CY20S and YES for 7 d at 25 °C in darkness. Two agar plugs (diameter 6 mm) were subsequently cut out from each medium and placed in an Eppendorf plastic vial and extracted with ethyl acetate / isopropanol (75:25, vol/vol) with 1 % formic acid. After ultrasonication for 50 mins the extraction liquid was transferred to another Eppendorf vial and the organic solvents evaporated. The chemical content was re-dissolved in 400 μl methanol and centrifuged at 13 400 rpm for 3 min. One μl liquid was injected into a HPLC-DAD with an additional fluorescence detector as described by Nielsen et al. (2011). For fluorescence detection, the excitation wavelength was 230 nm and the emission wavelengths were 333 nm and 450 nm. This allowed for sensitive detection of ochratoxins, aflatoxins, citrinin and indol alkaloids. Alkylphenone retention indices were calculated according to Frisvad and Thrane, 1987, Frisvad and Thrane, 1993.
Results and discussion
Phylogeny
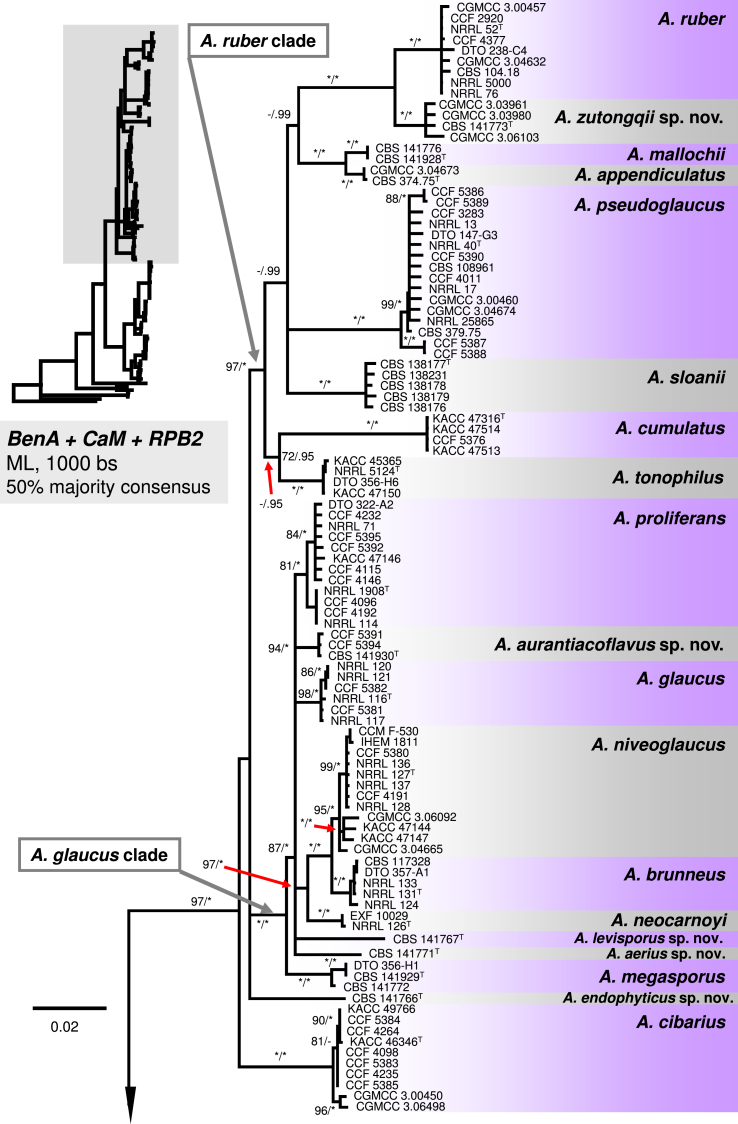
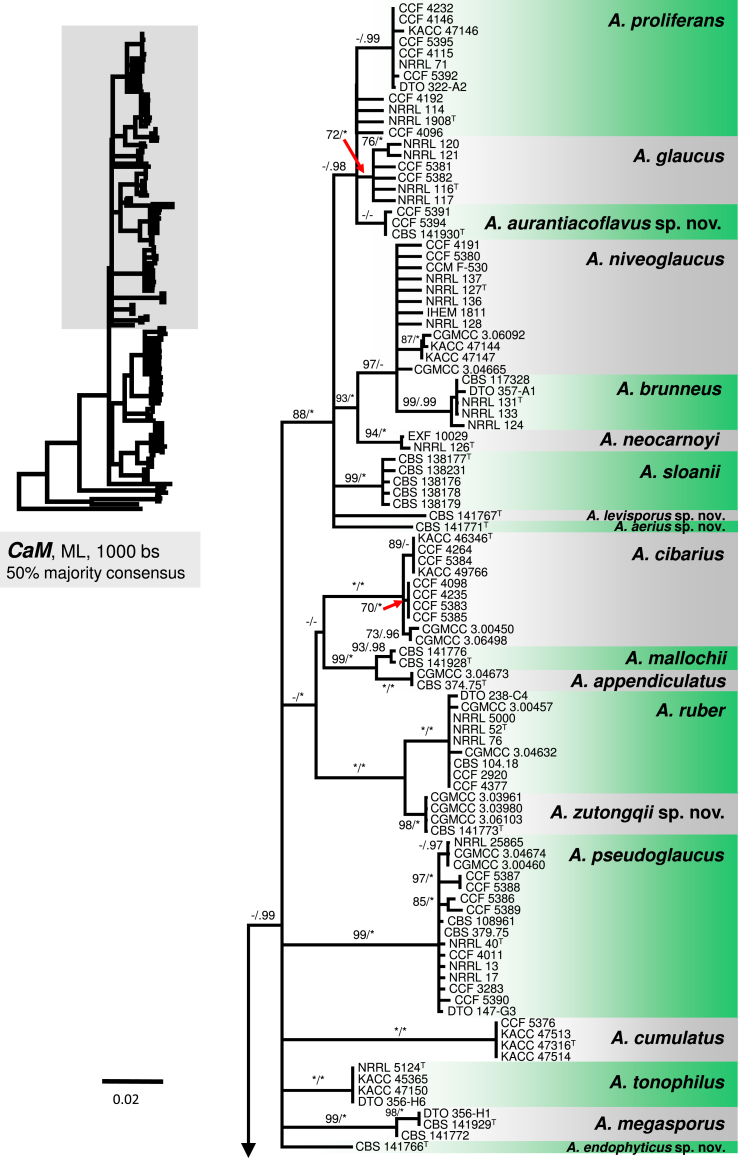
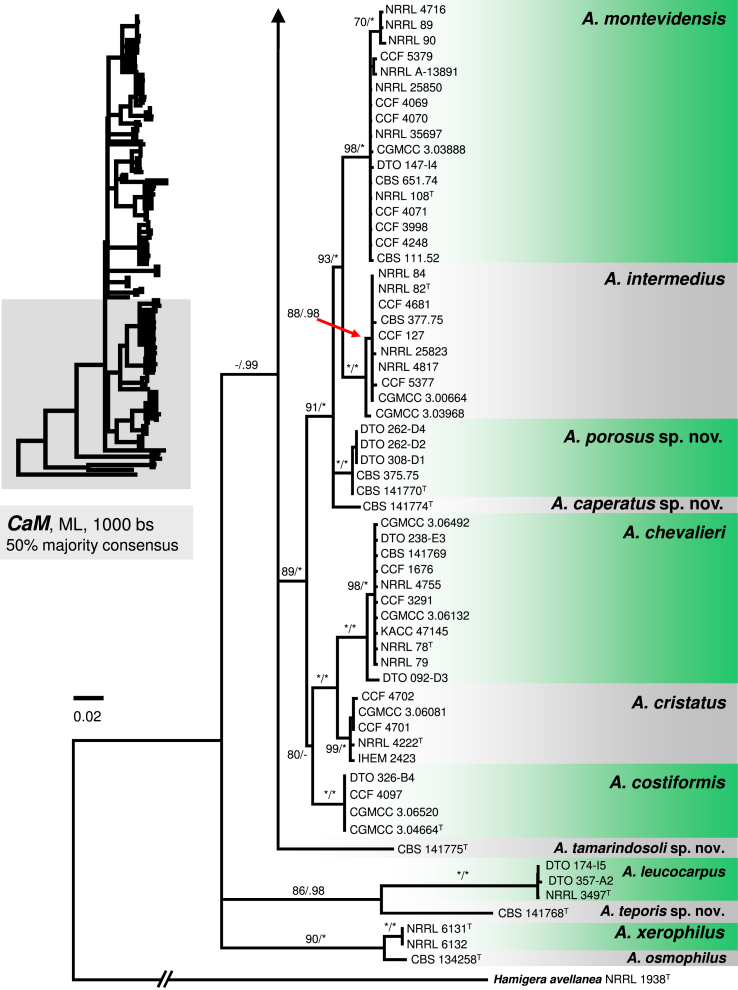
The phylogenetic relationships between 163 sect. Aspergillus strains were studied using concatenated sequence data of three loci: BenA, CaM and RPB2. In the 50 % majority consensus ML tree shown in Fig. 1, members of sect. Aspergillus are resolved in three major clades (named here the A. ruber, A. glaucus and A. chevalieri clades) and several, mostly basal, lineages containing one or two species. The Bayesian consensus tree was nearly identical to ML and therefore Bayesian posterior probabilities (PP) are shown on the ML tree nodes.
Fig. 1.
A 50 % majority rule Maximum likelihood consensus tree based on combined dataset of BenA, CaM and RPB2 sequences showing the relationship of species within Aspergillus sect. Aspergillus. Dataset contained 164 taxa, other alignment characteristics, partitioning scheme and nucleotide substitution models are listed in Table 3, Table 4. Maximum likelihood bootstrap proportion (bs) and Bayesian posterior probability (pp) are appended to nodes; only bs ≥ 70 % and pp ≥ 95 % are shown, lower supports are indicated with a hyphen, whereas asterisks indicate full support (100 % bs or 1.00 pp); ex-type strains are designated by a superscript T. The tree is rooted with Hamigera avellanea NRRL 1938T.
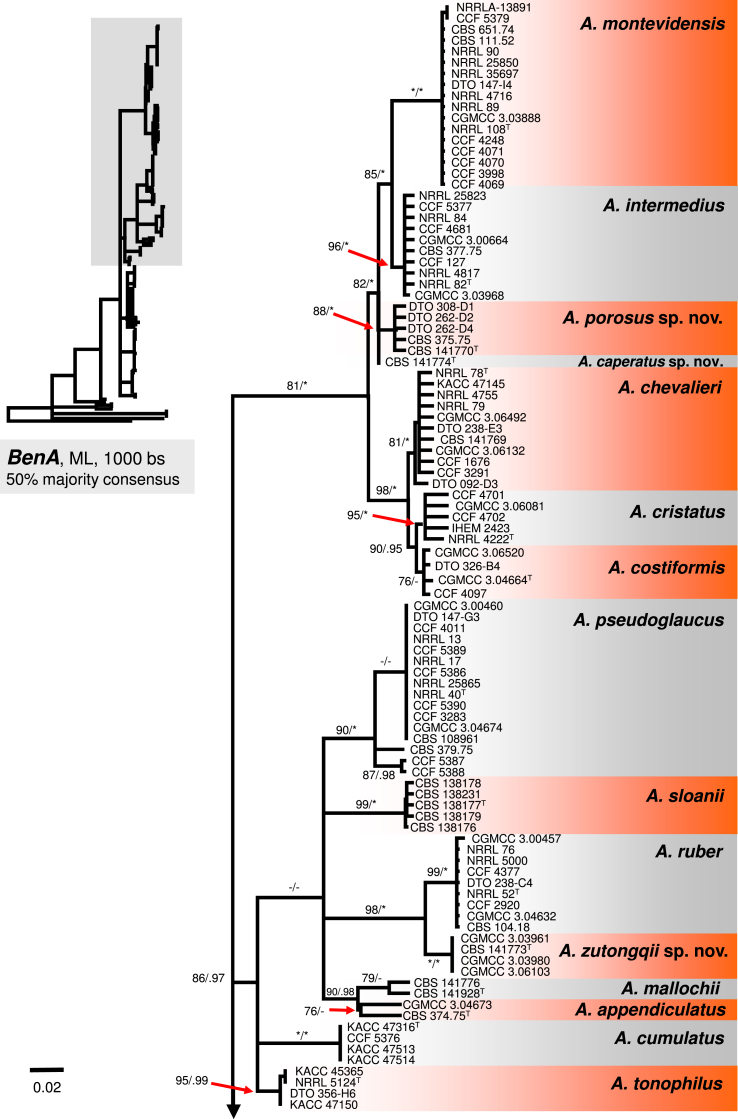
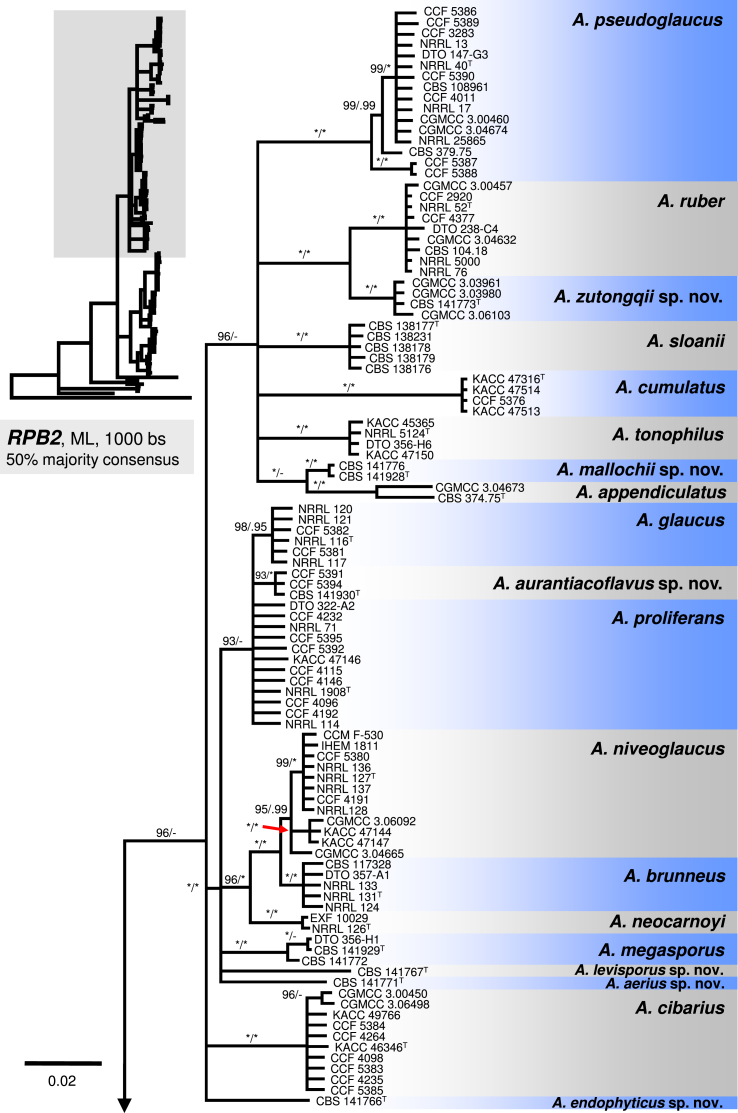
The A. ruber clade contains A. appendiculatus, A. cumulatus, A. mallochii, A. pseudoglaucus, A. ruber, A. sloanii, A. tonophilus, and a new species A. zutongqii, with four strains originating from China (CBS 141773, CGMCC 3.03961, CGMCC 3.03980, CGMCC 3.06103) forming a sister clade to A. ruber. Its placement as sister to A. ruber is further supported by the single-gene trees (Fig. 2, Fig. 3, Fig. 4). Aspergillus fimicola (ex-type: CBS 101747), Aspergillus glaber (ex-type: CBS 379.75) and A. reptans (ex-type: NRRL 13) resolve in the A. pseudoglaucus lineage; A. tuberculatus (ex-type: CBS 101748) and A. athecius (ex-type: NRRL 5000) in the A. ruber lineage; and A. aridicola (ex-type: CBS 101746) in the A. appendiculatus lineage. Very similar topologies of the A. ruber clade was produced by phylogenetic analyses based on BenA (86 % BS / 0.97 PP; Fig. 2) and RPB2 (96 % BS / 0.79 PP; Fig. 4), while these topologies were not supported by CaM-based phylogeny (Fig. 3). All eight lineages within the clade were strongly supported in the combined phylogenetic analyses as well as single-gene analyses (BS ≥ 90 %, PP ≥ 0.98). The exception is in the BenA phylogeny where nodes bearing A. mallochii and A. appendiculatus had limited support (79 % BS / 0.92 PP and 76 % BS / 0.88 PP, respectively; Fig. 2).
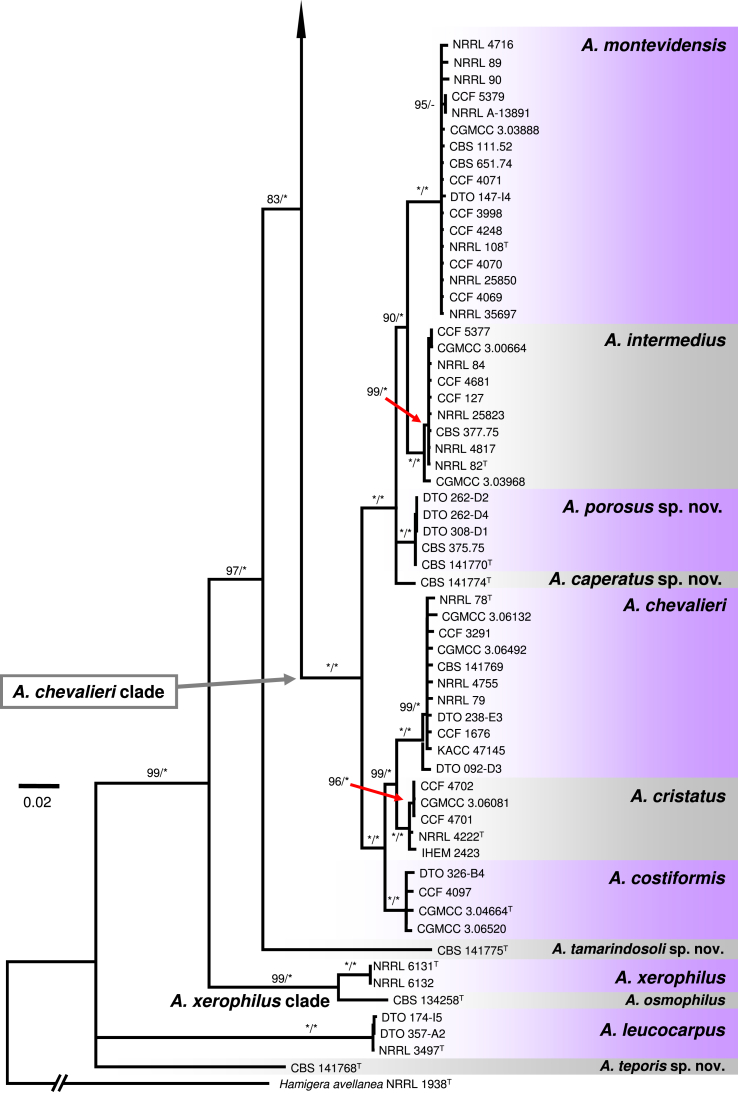
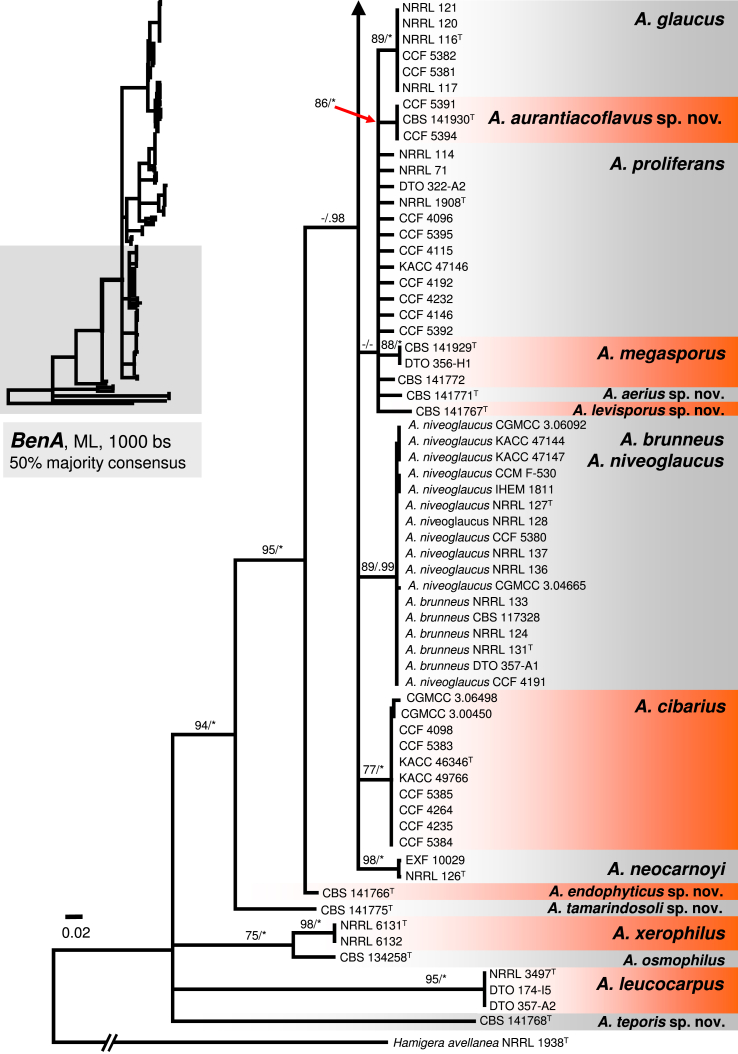
Fig. 2.
A 50 % majority rule Maximum likelihood consensus tree based on partial β-tubulin (BenA) sequences showing the relationship of species within Aspergillus sect. Aspergillus. Maximum likelihood bootstrap proportion (bs) and Bayesian posterior probability (pp) are appended to nodes; only bs ≥ 70 % and pp ≥ 95 % are shown, lower supports are indicated with a hyphen, whereas asterisks indicate full support (100 % bs or 1.00 pp); ex-type strains are designated by a superscript T. The tree is rooted with Hamigera avellanea NRRL 1938T.
Fig. 3.
A 50 % majority rule Maximum likelihood consensus tree based on partial calmodulin (CaM) sequences showing the relationship of species within Aspergillus sect. Aspergillus. Maximum likelihood bootstrap proportion (bs) and Bayesian posterior probability (pp) are appended to nodes; only bs ≥ 70 % and pp ≥ 95 % are shown, lower supports are indicated with a hyphen, whereas asterisks indicate full support (100 % bs or 1.00 pp); ex-type strains are designated by a superscript T. The tree is rooted with Hamigera avellanea NRRL 1938T.
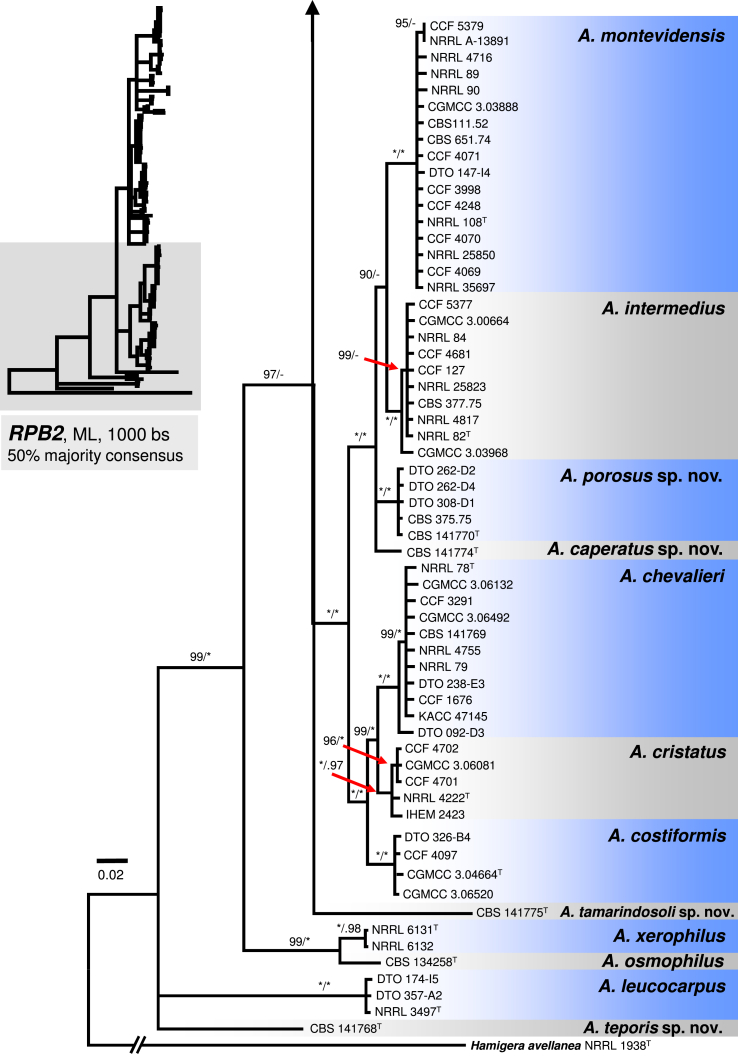
Fig. 4.
A 50 % majority rule Maximum likelihood consensus tree based on partial RNA polymerase II second largest subunit (RPB2) sequences showing the relationship of species within Aspergillus sect. Aspergillus. Maximum likelihood bootstrap proportion (bs) and Bayesian posterior probability (pp) are appended to nodes; only bs ≥ 70 % and pp ≥ 95 % are shown, lower supports are indicated with a hyphen, whereas asterisks indicate full support (100 % bs or 1.00 pp); ex-type strains are designated by a superscript T. The tree is rooted with Hamigera avellanea NRRL 1938T.
The A. glaucus clade contains A. brunneus, A. glaucus, A. megasporus, A. niveoglaucus, A. neocarnoyi, A. proliferans and three new species (Fig. 1). Three isolates originating from the USA (CBS 141930, CCF 5391 and CCF 5394) formed a well-supported clade (94 % BS / 1.00 PP) closely related to A. glaucus and A. proliferans (Fig. 1); this clade showed moderate to high support in BenA and RPB2 phylogenies (Fig. 2, Fig. 4), but weak support in the CaM tree (Fig. 3). The clade is introduced as a new species, A. aurantiacoflavus, in the taxonomy section. All three species (A. glaucus, A. proliferans and A. aurantiacoflavus) have fixed single-nucleotide polymorphisms at BenA, CaM and RPB2 loci that guarantee their reliable discrimination from each other (positions in particular alignments available in Dryad Digital Repository — BenA: positions 53, 138, 219 and 297; CaM: positions 2, 131, 460, 600; RPB2: positions 211, 279, 666). CBS 141771 and CBS 141767 formed distinct single-isolate lineages nested in the A. glaucus clade but with unresolved position. They are distantly related to each other and remaining taxa in the clade based on CaM and RPB2 data, and are proposed below as new species A. aerius and A. levisporus. In the BenA phylogeny, these two species are resolved on a weakly supported branch with A. proliferans, A. glaucus, A. aurantiacoflavus and A. megasporus (Fig. 2), but their sequences contain numerous substitutions sufficient for reliable identification. Aspergillus medius (ex-type: NRRL 124) belongs in the A. brunneus lineage, A. parviverruculosus (ex-type: CBS 101750) in the A. niveoglaucus lineage, and A. manginii (ex-type: NRRL 117) and A. umbrosus (ex-type: NRRL 120) in the A. glaucus lineage. The A. proliferans lineage includes a strictly anamorphic ex-type strain NRRL 1908 and numerous isolates producing eurotium-like sexual state. Tree topologies of the A. glaucus clade in the CaM, RPB2 and combined trees are nearly identical (Fig. 1, Fig. 3, Fig. 4). In contrast, the BenA locus has only limited discriminatory power in this clade and many species were collapsed in a polytomy (Fig. 2). But still BenA sequences are sufficient for identification of all species except A. brunneus and A. niveoglaucus. Species represented by at least two strains usually gained high or moderate support in ML and BI analyses based on combined data, or CaM and RPB2 genes (Fig. 1, Fig. 3, Fig. 4) except A. aurantiacoflavus, A. glaucus and A. proliferans that were weakly supported in single-gene phylogenies. However, recognition of these species is supported by phenotype, especially by morphology of ascospores (see below). On the other hand, additional strongly supported clades delimited by same analyses (Fig. 1, Fig. 3, Fig. 4) within A. glaucus, A. niveoglaucus and A. proliferans lineages, had no or very limited phenotypic support, which is the reason why we decided for broader species concept rather than for splitting these species.
The A. chevalieri clade includes A. chevalieri, A. costiformis, A. cristatus, A. intermedius, A. montevidensis and two new species A. caperatus and A. porosus. Aspergillus caperatus is represented by CBS 141774 from South Africa. Aspergillus porosus is represented by five strains originating from Turkey and Israel (CBS 141770, CBS 375.75, DTO 308-D1, DTO 262-D2, DTO 262-D4), they formed a clade with full support that is related to A. caperatus, A. intermedius and A. montevidensis (Fig. 1). The topology of this subclade is identical in single-gene phylogenies and all lineages are strongly supported (Fig. 2, Fig. 3, Fig. 4). The rest three species in the A. chevalieri clade cluster together and form another strongly supported subclade (Fig. 1) with highly congruent topology shown in single-gene phylogenies (Fig. 2, Fig. 3, Fig. 4). Aspergillus spiculosus (ex-type: CBS 377.75) is included in the A. intermedius lineage; A. hollandicus (ex-type: CBS 518.65), A. heterocaryoticus (ex-type: CBS 410.65) and A. vitis (ex-type: CBS 651.74) in the A. montevidensis lineage.
The remaining taxa belonging to sect. Aspergillus were distantly related to species from these three main clades and formed remote lineages with not fully resolved or basal position in the trees. Aspergillus cibarius and strain CBS 141766 (described below as A. endophyticus) were resolved in a basal position adjacent to A. ruber and A. glaucus clades (Fig. 1); their position varies between single-gene trees (Fig. 2, Fig. 3, Fig. 4). Aspergillus xerophilus and closely related A. osmophilus form A. xerophilus clade, A. leucocarpus, A. tamarindosoli and A. teporis formed basal lineages distantly related to core species of sect. Aspergillus (Fig. 1, Fig. 2, Fig. 3, Fig. 4).
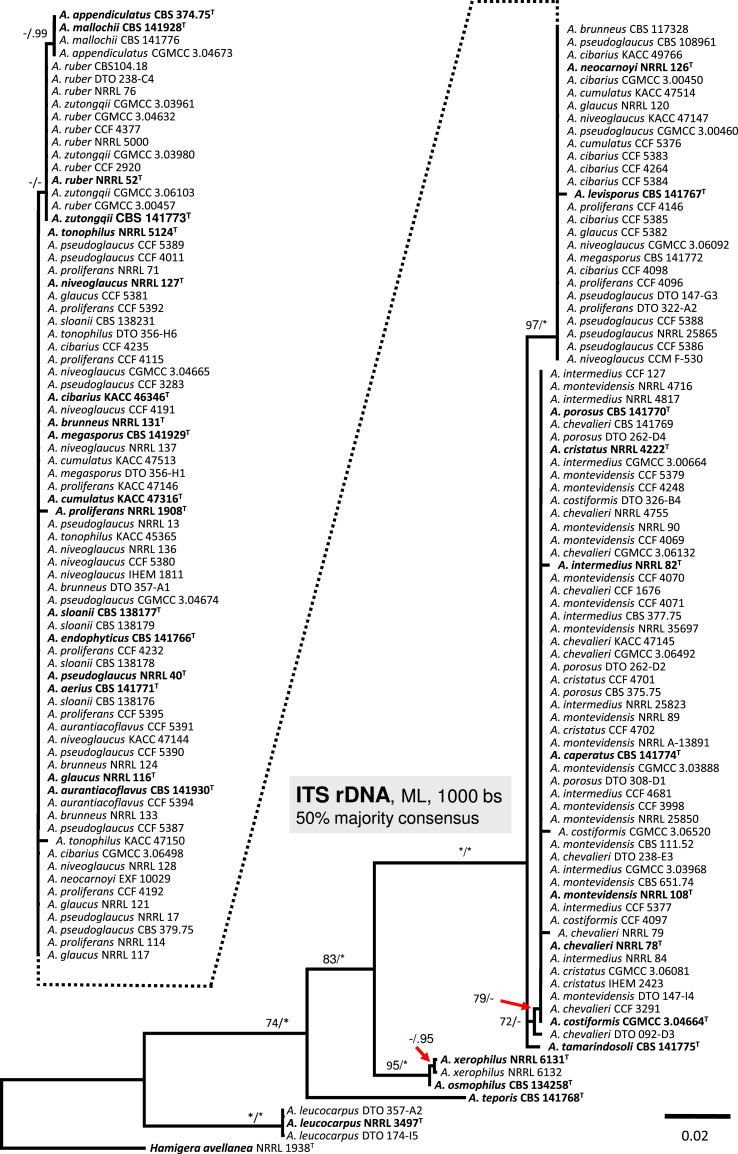
ITS sequences do not contain sufficient variation for distinguishing among sect. Aspergillus species (Fig. 5), and therefore this locus was excluded from the combined phylogenetic analysis. Only five species had unique ITS sequences (A. tamarindosoli, A. xerophilus, A. osmophiluc, A. leucocarpus and A. teporis; Fig. 5); identical sequence is shared for species from the A. chevalieri clade (n = 7); A. appendiculatus and A. mallochii; and A. ruber and A. zutongqii. All remaining species (n = 15) are indistinguishable by ITS sequences. Intraspecific single-nucleotide polymorphisms were observed in sequences of A. proliferans, A. tonophilus, A. intermedius, A. costiformis and A. chevalieri.
Fig. 5.
A 50 % majority rule Maximum likelihood consensus tree based on ITS sequences. Maximum likelihood bootstrap proportion (bs) and Bayesian posterior probability (pp) are appended to nodes; only bs ≥ 70 % and pp ≥ 95 % are shown, lower supports are indicated with a hyphen, whereas asterisks indicate full support (100 % bs or 1.00 pp); ex-type strains are designated by a superscript T. The tree is rooted with Hamigera avellanea NRRL 1938T.
Peterson (2008) accepted 15 species in sect. Aspergillus based on congruence analysis of BenA, CaM, ID and RPB2. Fourteen sexual species were placed under the Eurotium name, the only anamorphic species A. proliferans formed a monophylic group with two ascomata producing strains identified as “Eurotium rubrum” and “E. mangini” (NRRL 71 and NRRL 114). The phylogenetic identity of anamorphic ex-type strain NRRL 1908 and other ascosporic strains was additionally supported by Hubka et al. (2012) and Asgari et al. (2014). Hubka et al. (2013a) applied the GCPSR criteria in sect. Aspergillus based on the same four loci and adopted Aspergillus names for Eurotium species. In their study, 17 species were accepted, all of which can be distinguished by CaM or RPB2 loci, and the concept of A. proliferans was extended by a description of its sexual state. In this study, 31 well-supported phylogenetic lineages representing species are recognized. This conclusion is based on phylogenetic analysis of concatenated and partitioned sequence data, comparison of topologies of single-gene phylogenetic trees and reflection of phenotypic data (see below). All species can be distinguished by CaM or RPB2 sequences, while BenA can be used to identify 29 species, with A. brunneus and A. niveoglaucus sharing identical BenA sequences.
Morphology and physiology
Members of sect. Aspergillus are generally characterized by yellow cleistothecia (the only exception is A. leucocarpus, which produces white cleistothecia), lenticular, hyaline ascospores, uniseriate conidiophore heads and globose, subglobose or ellipsoidal conidia. In the past, colony appearance, ascospore and conidial morphology were emphasized to differentiate species in this section (Thom and Raper, 1941, Thom and Raper, 1945, Raper and Fennell, 1965, Blaser, 1975, Kozakiewicz, 1989, Sun and Qi, 1994, Guarro et al., 2012). This led to many species recognized which do not necessarily correspond to species based on recent phylogenetic data.
Macromorphology
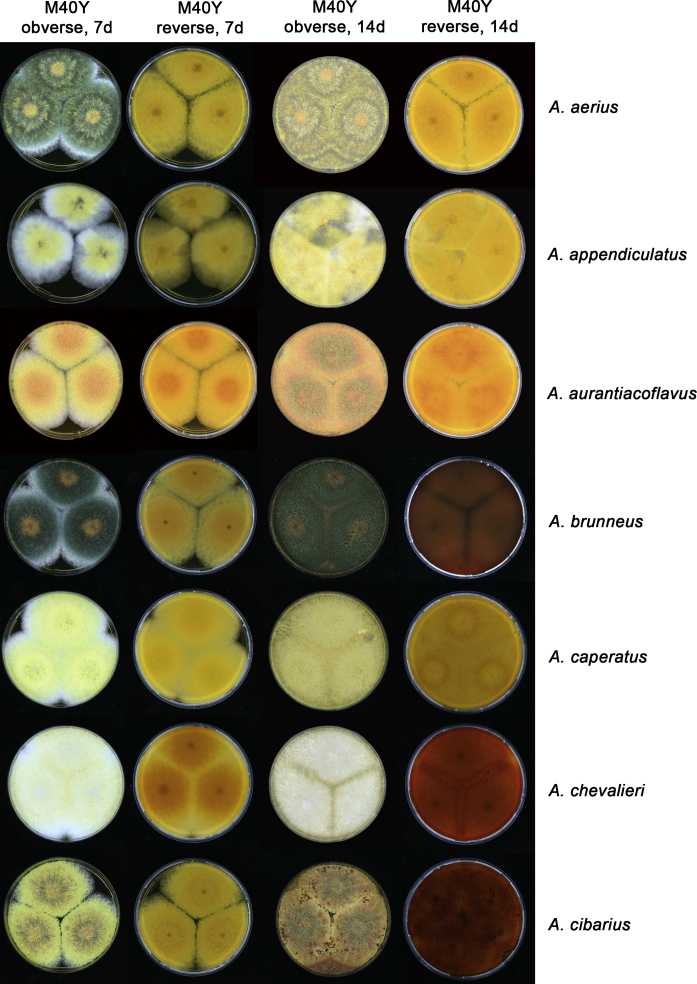
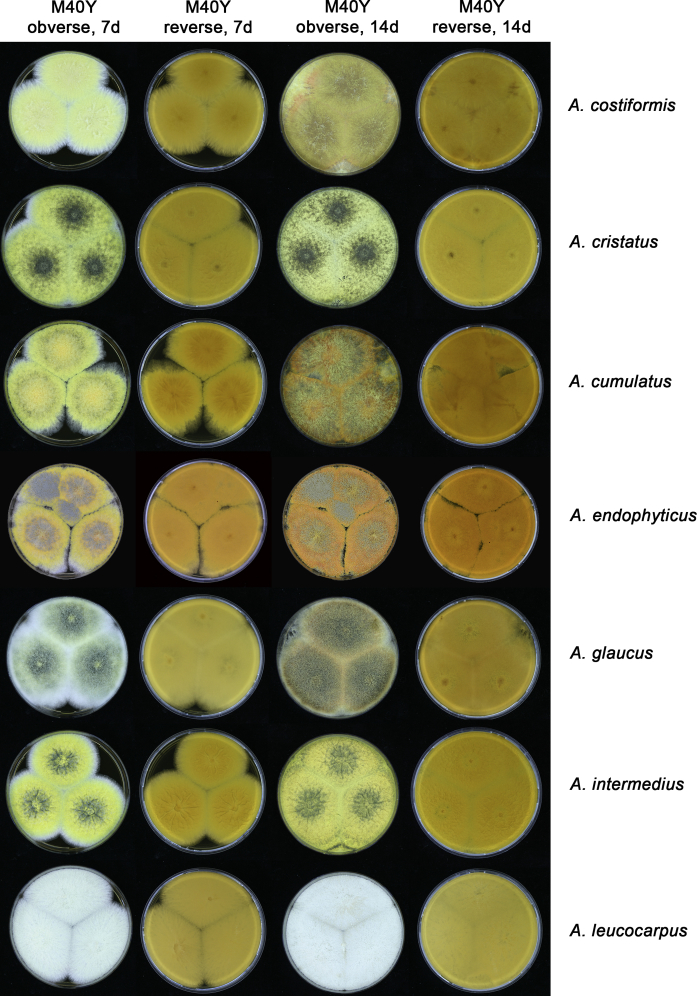
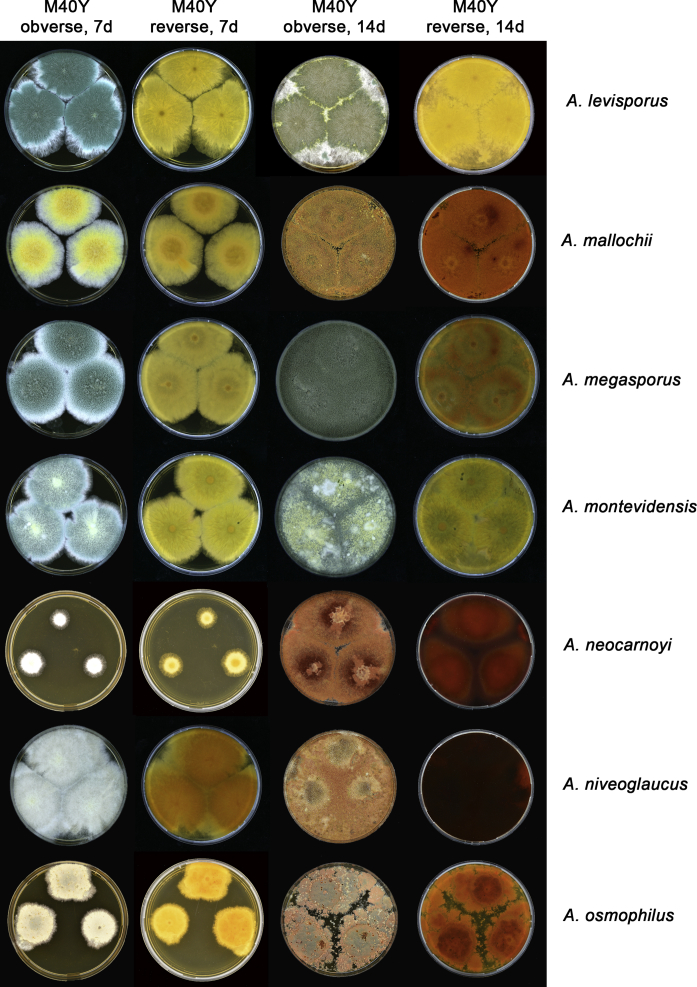
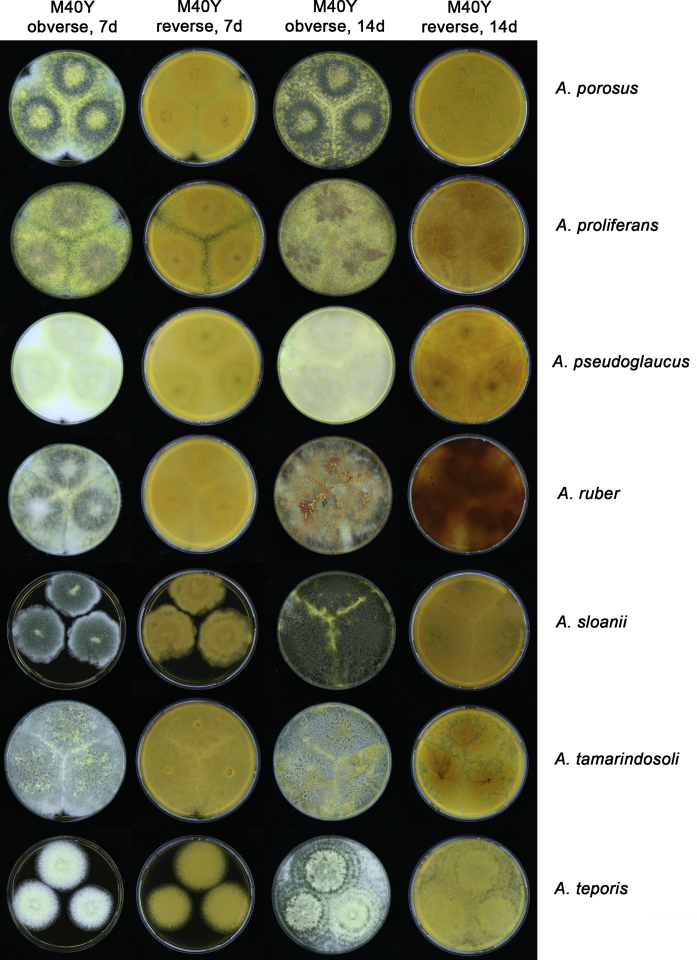
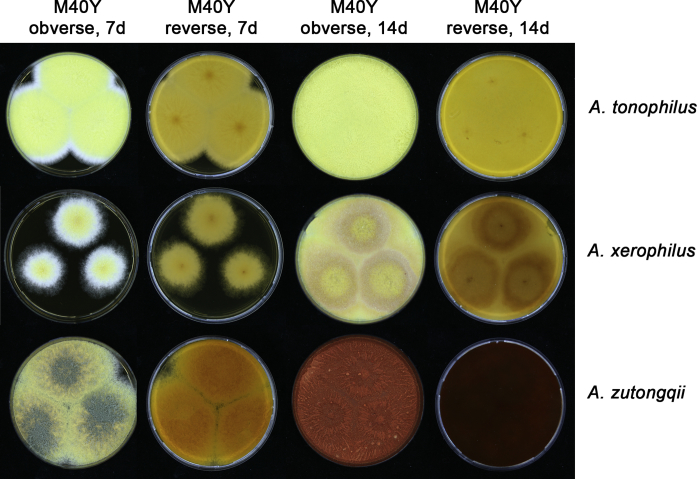
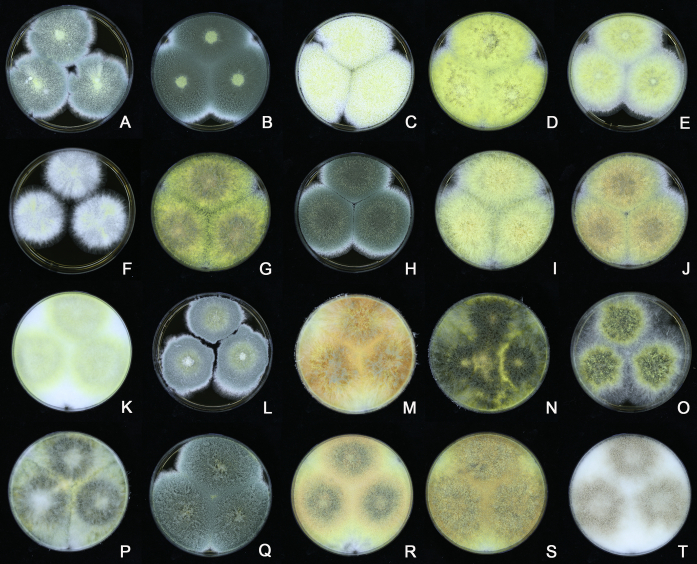
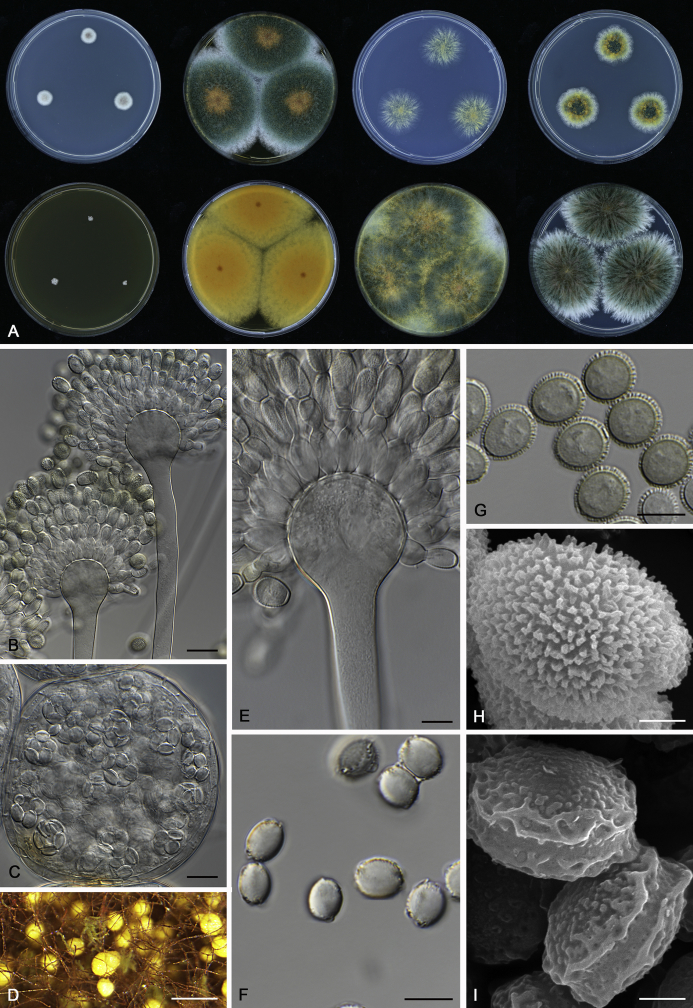
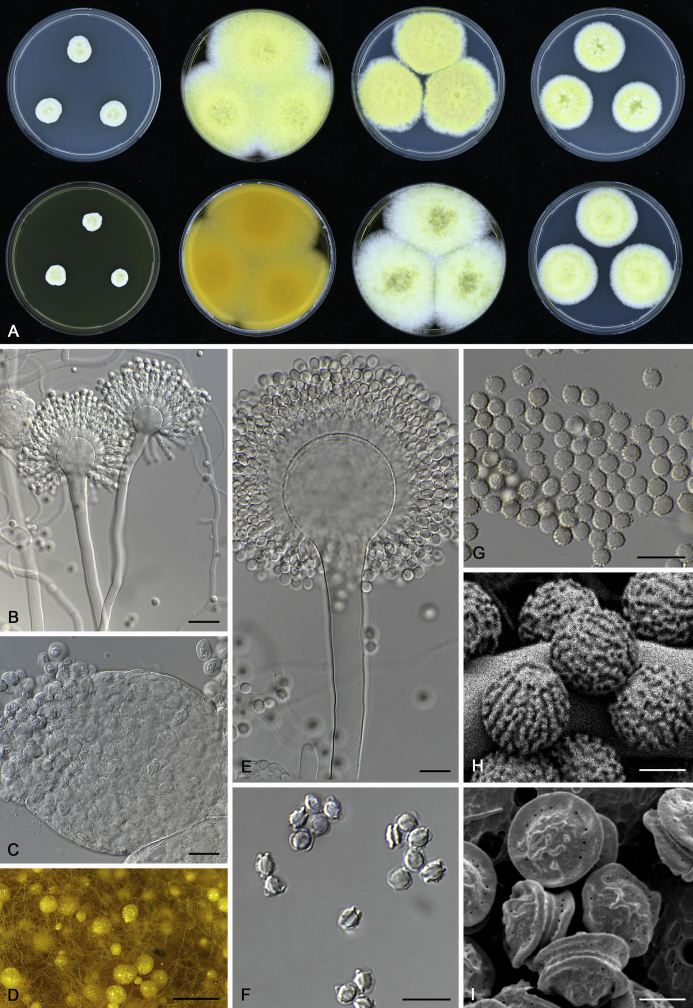
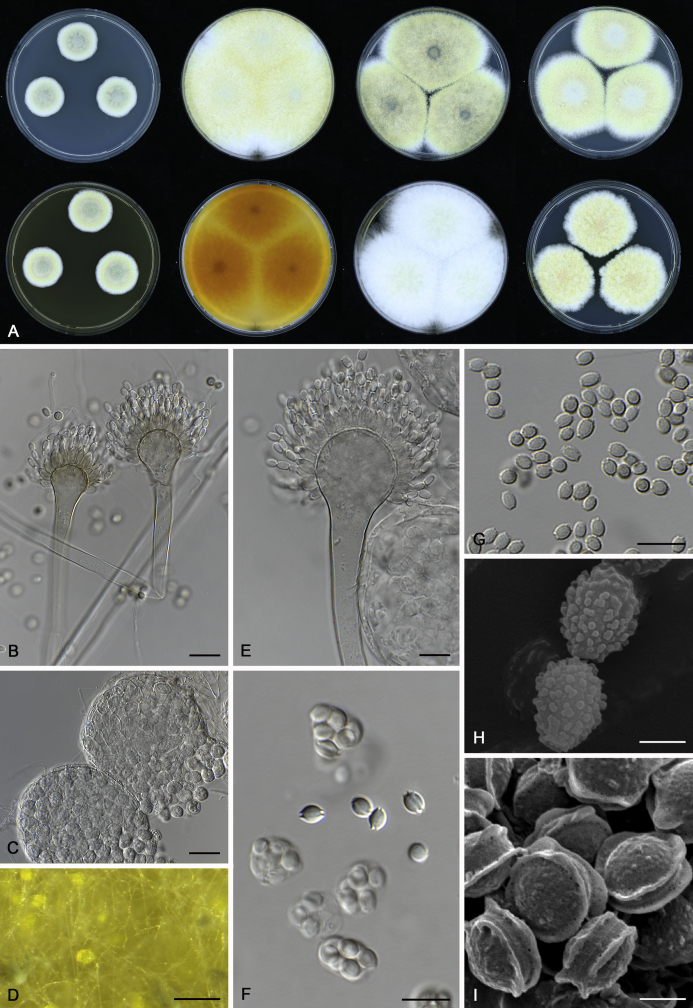
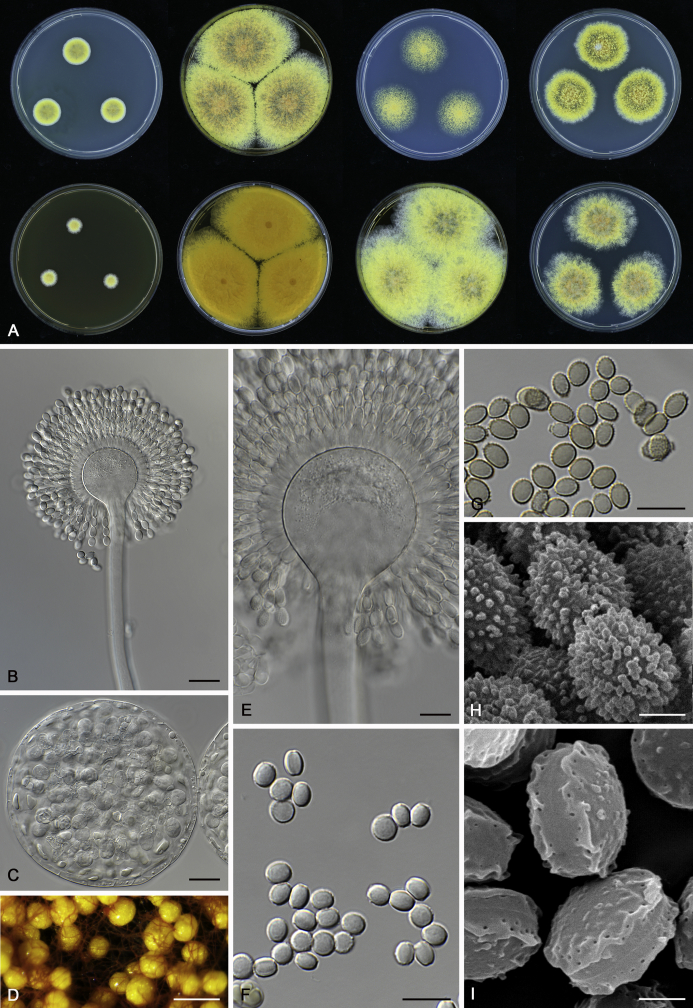
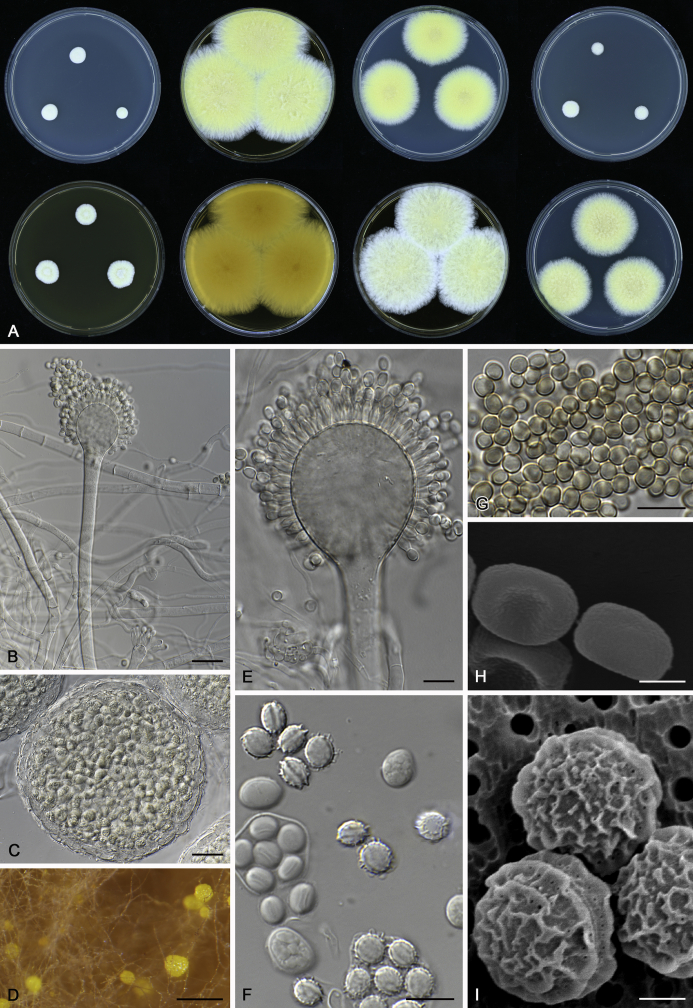
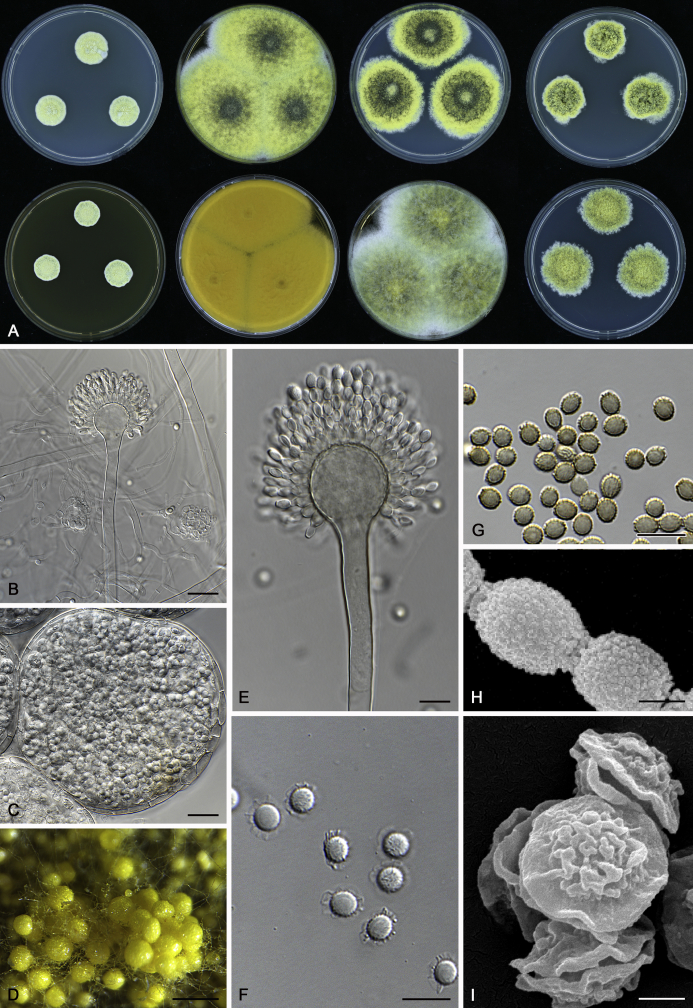
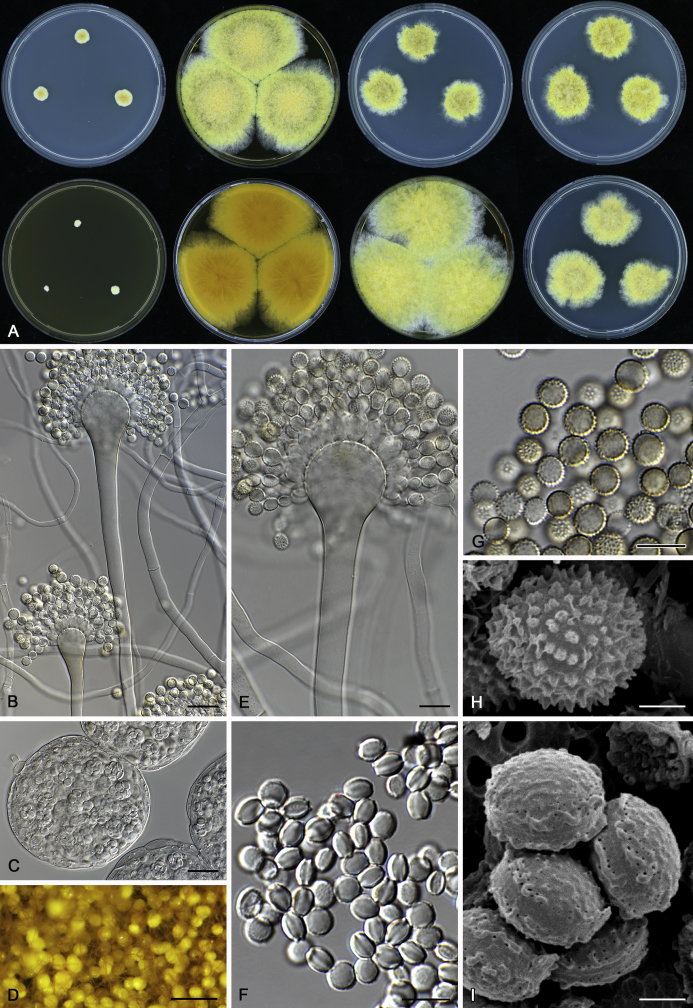
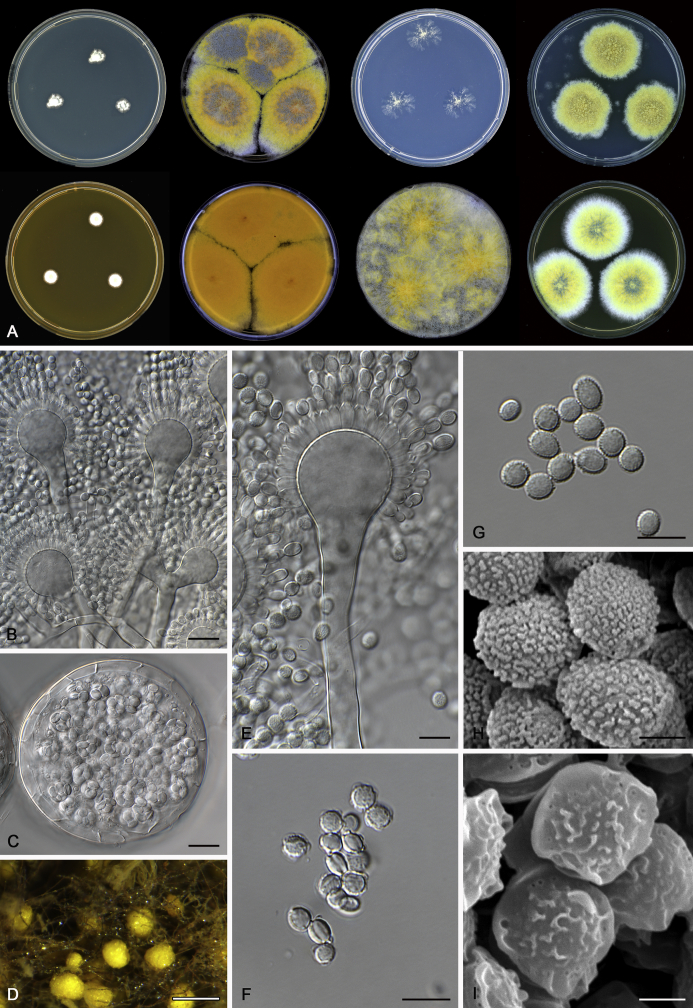
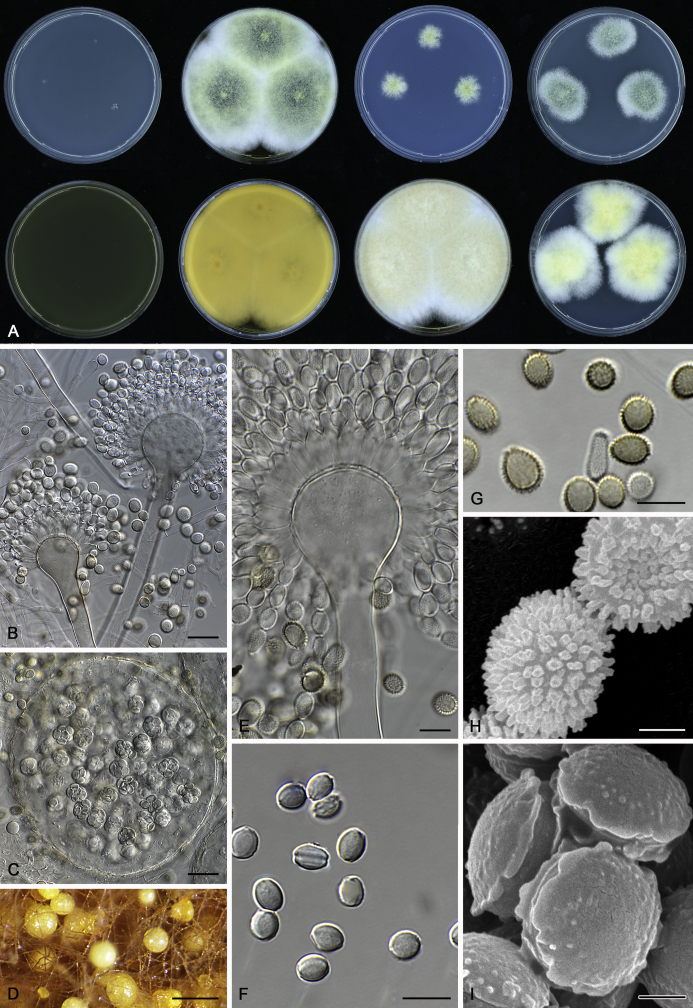
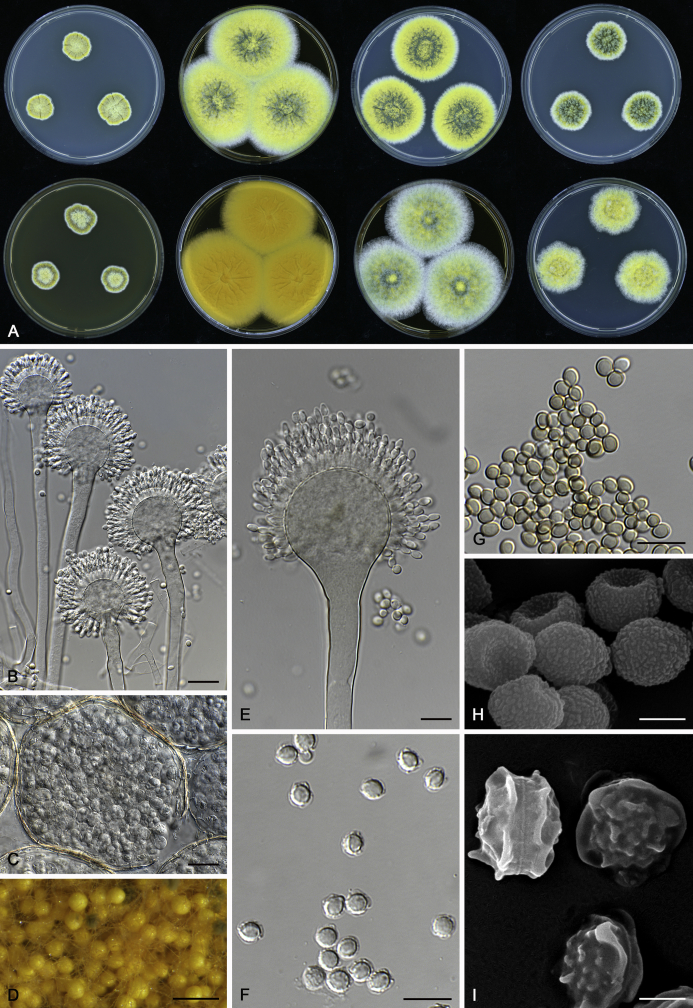
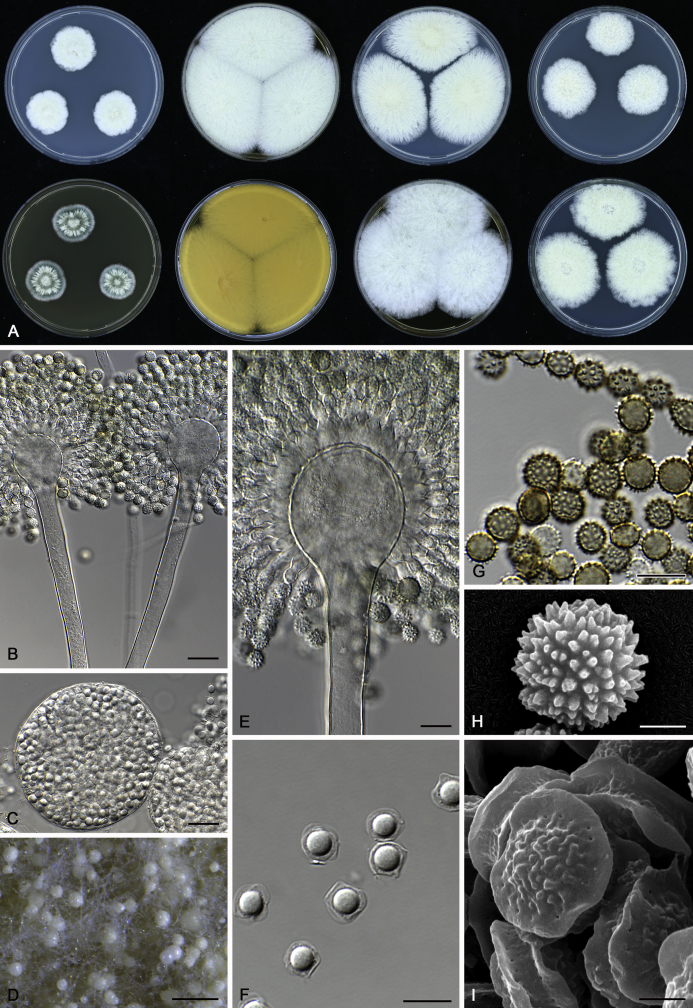
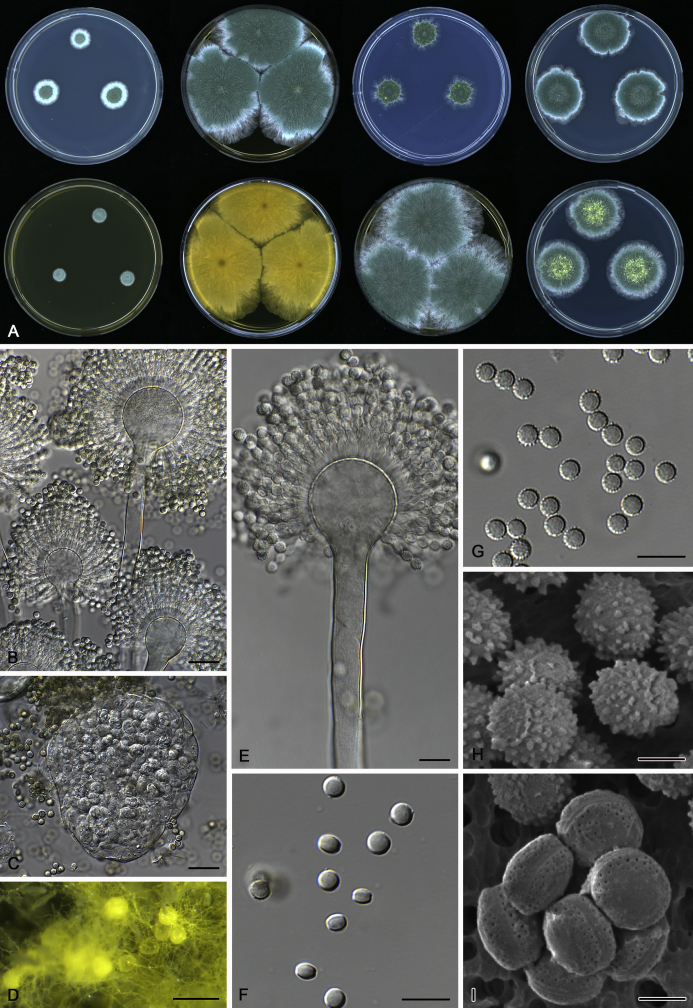
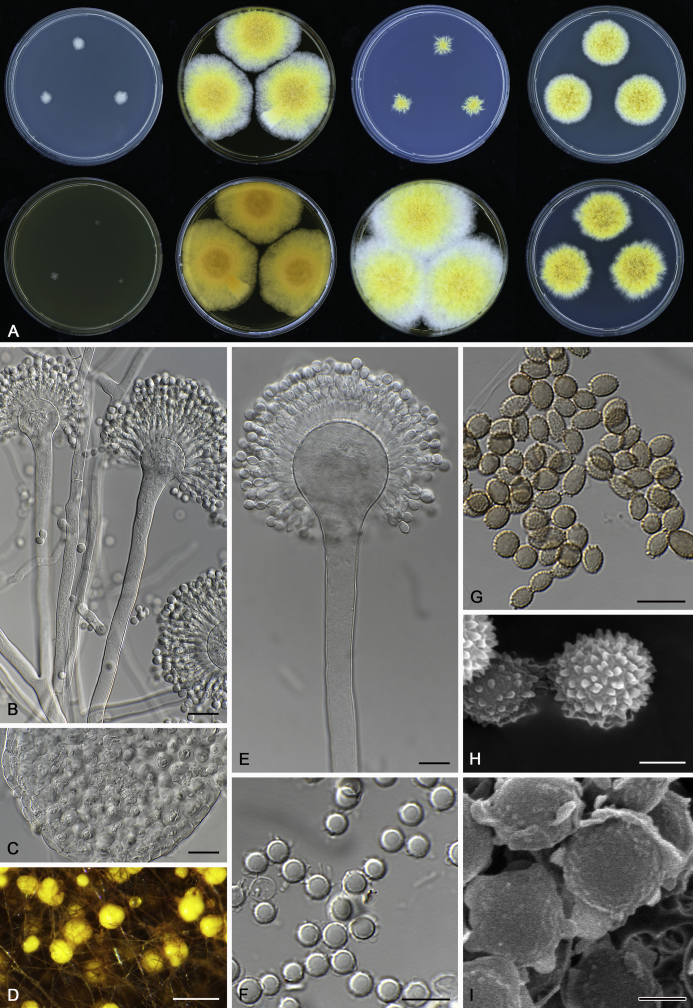
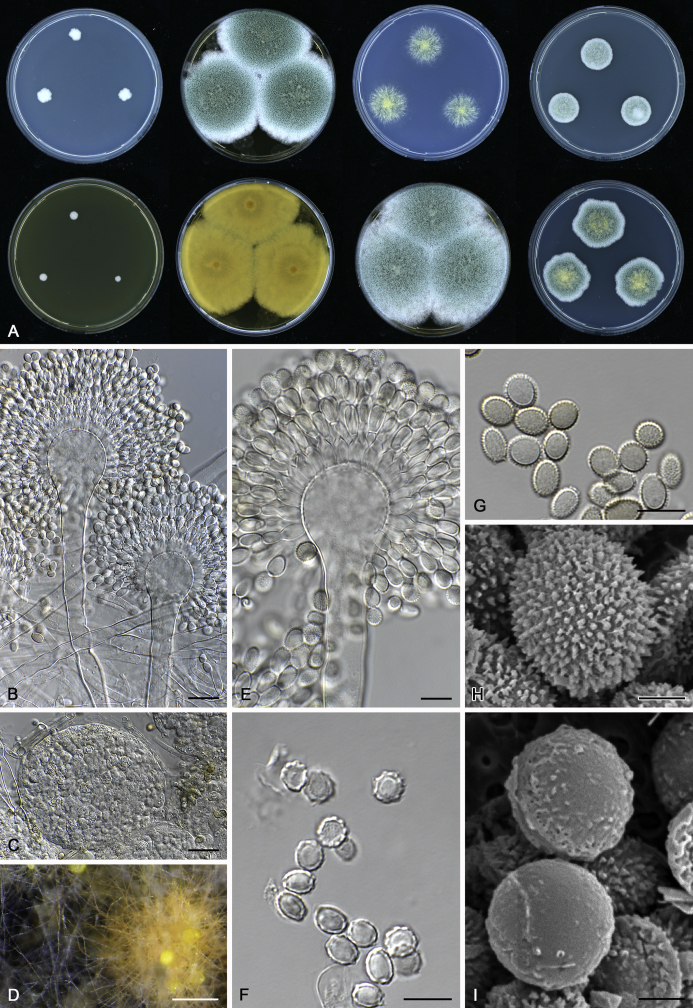
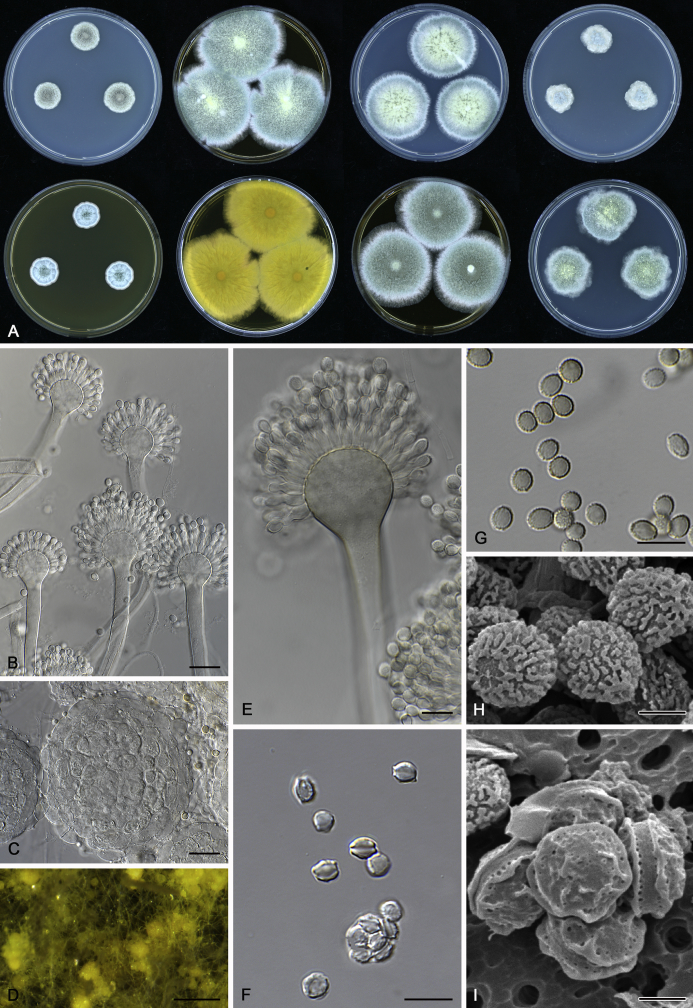
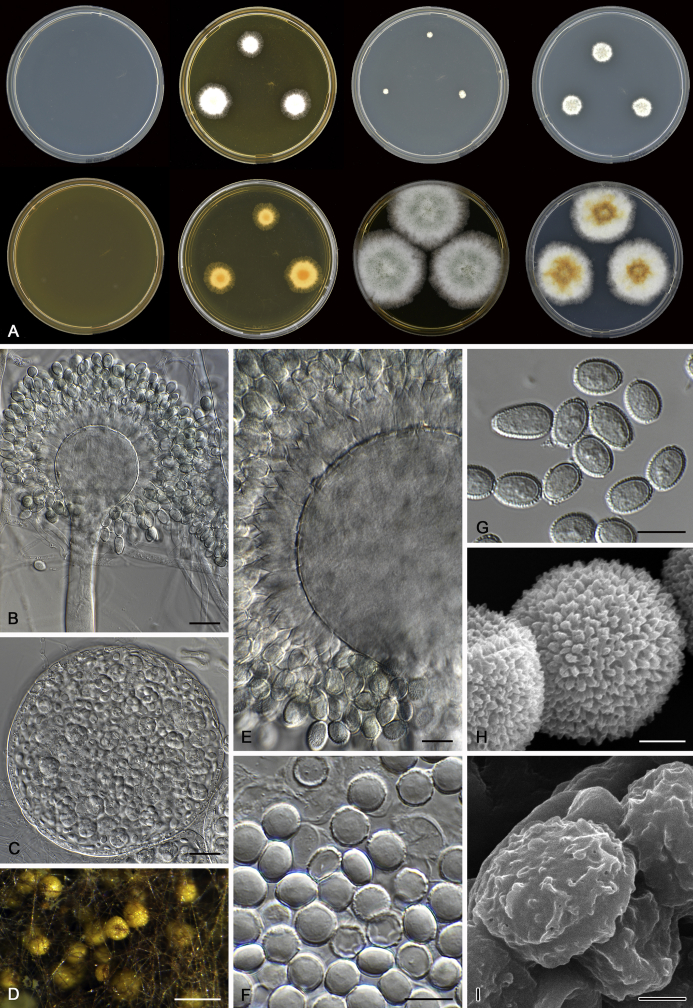
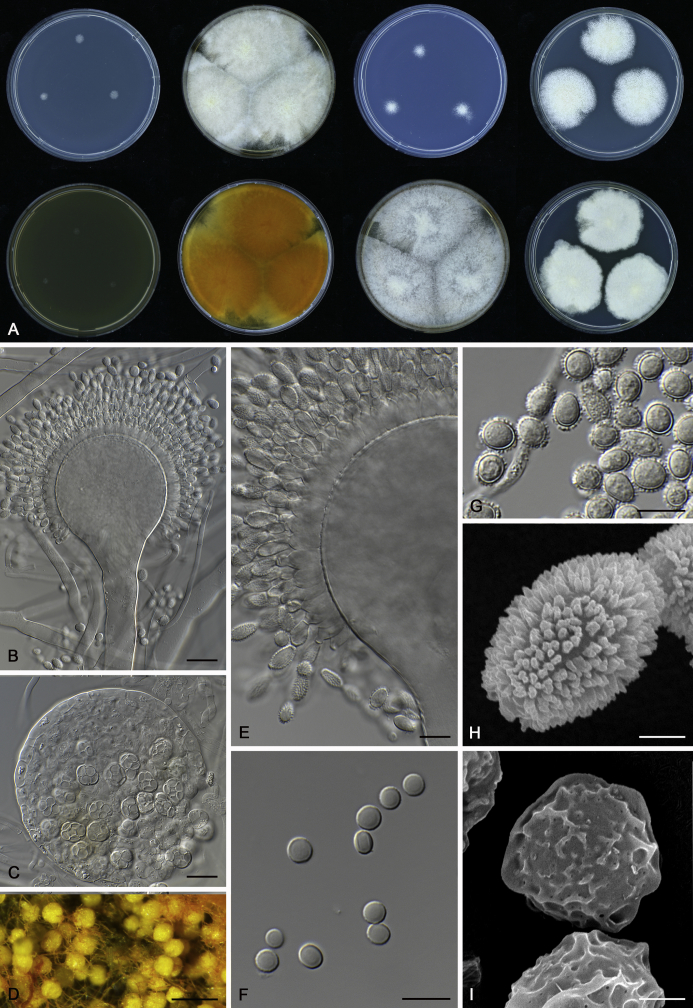
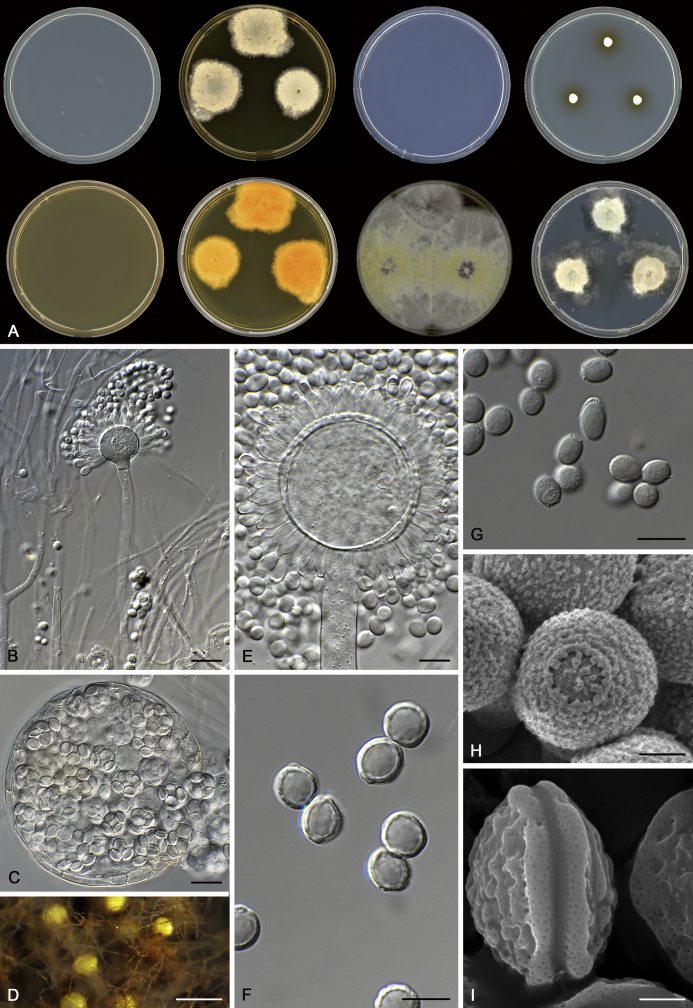
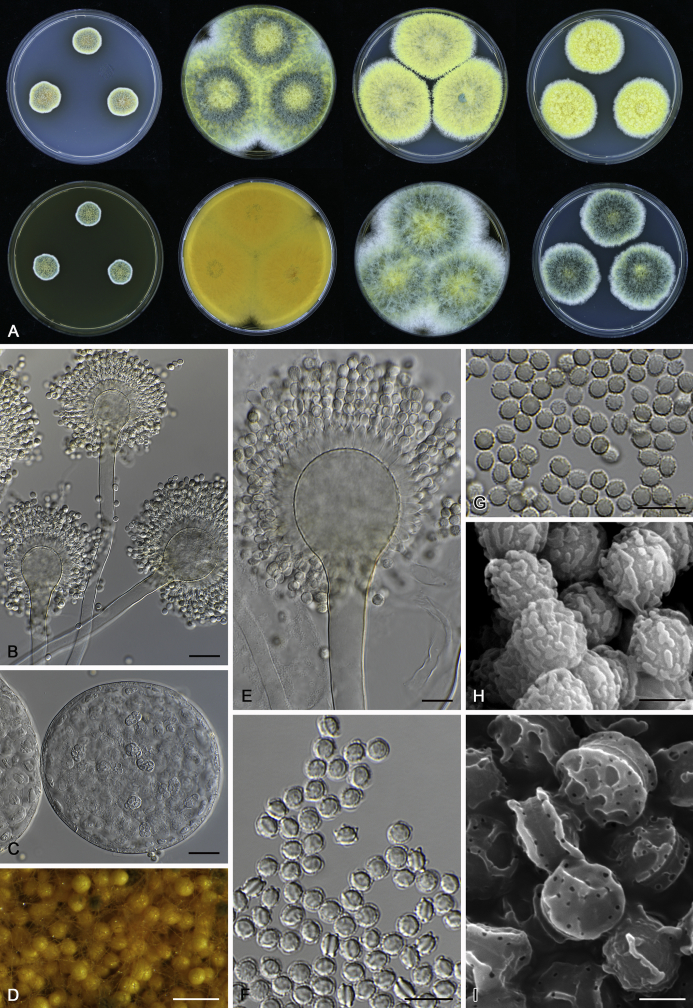
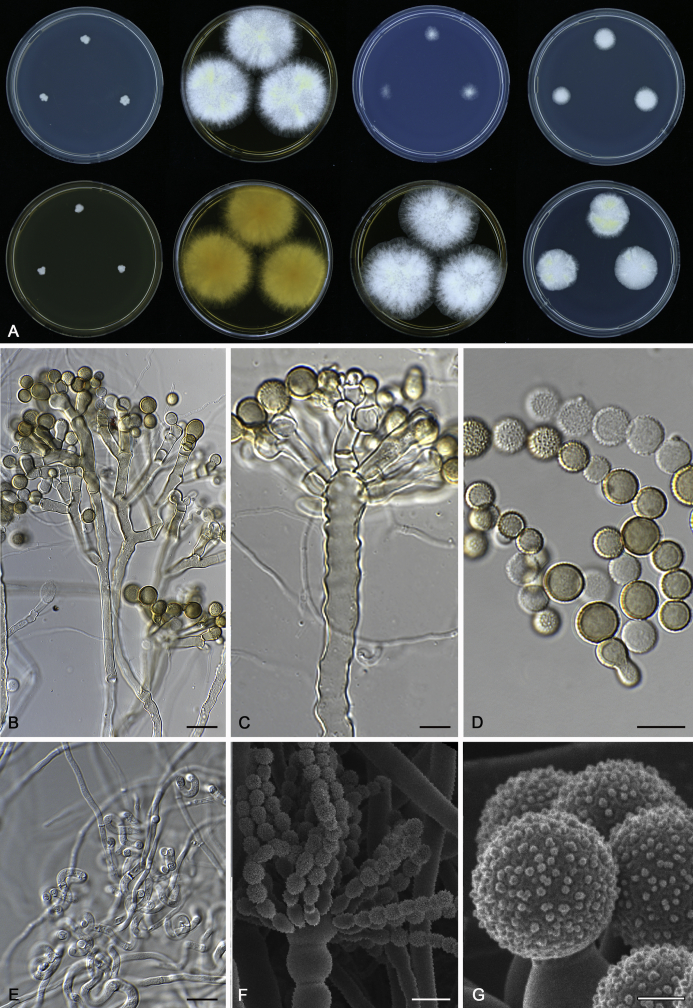
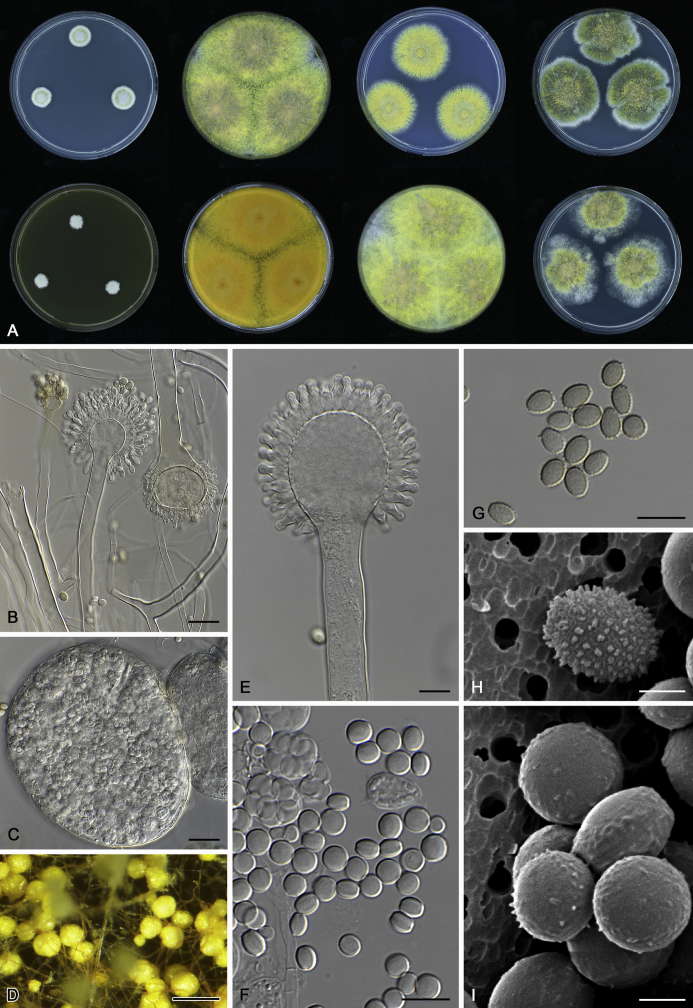
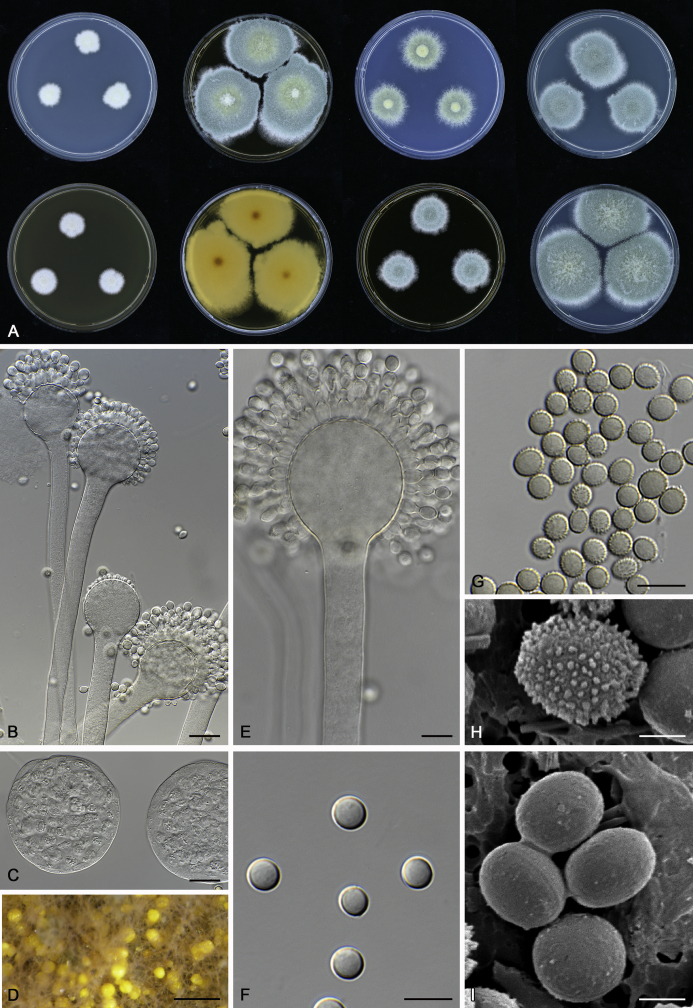
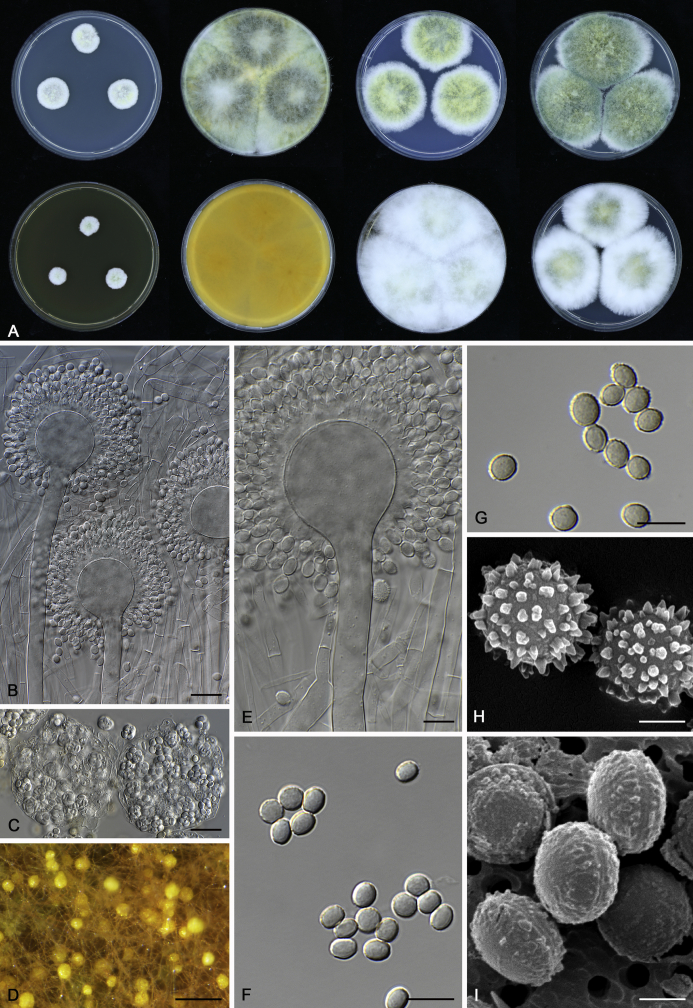
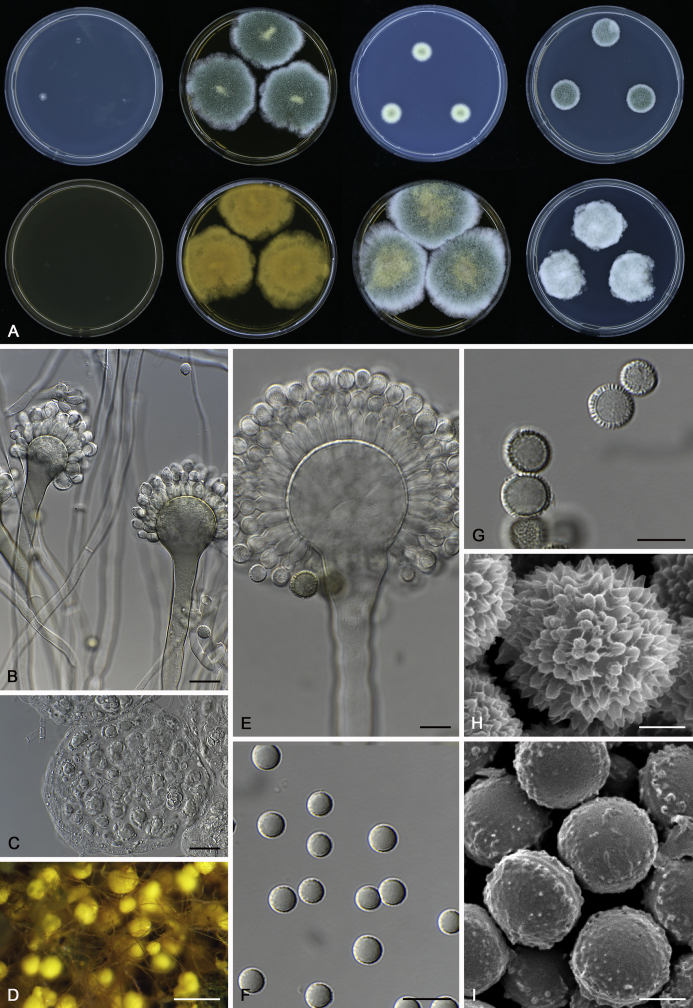
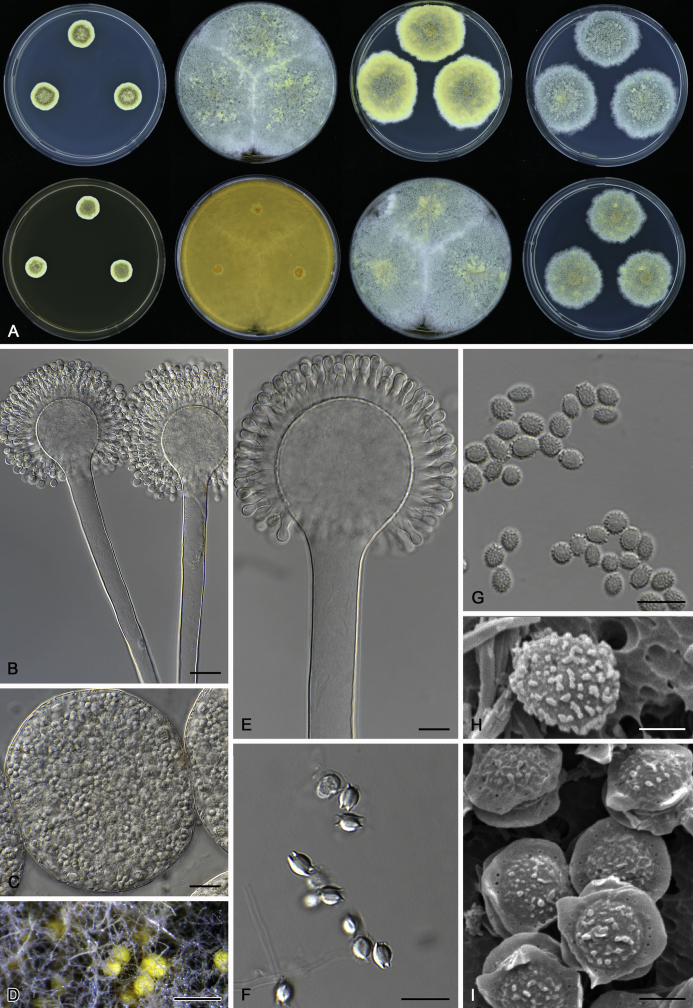
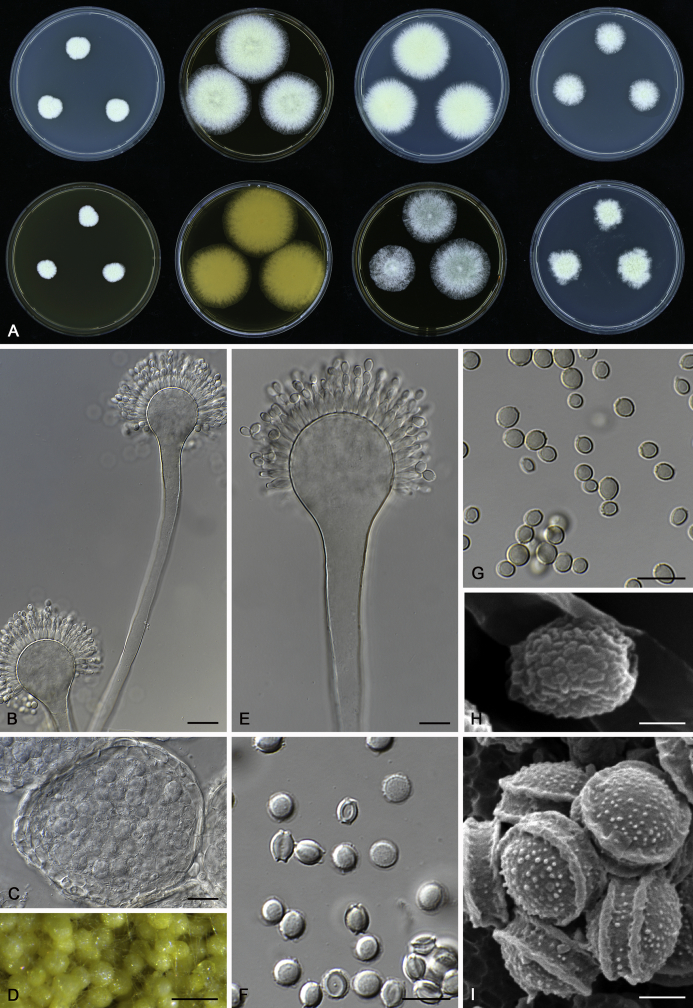
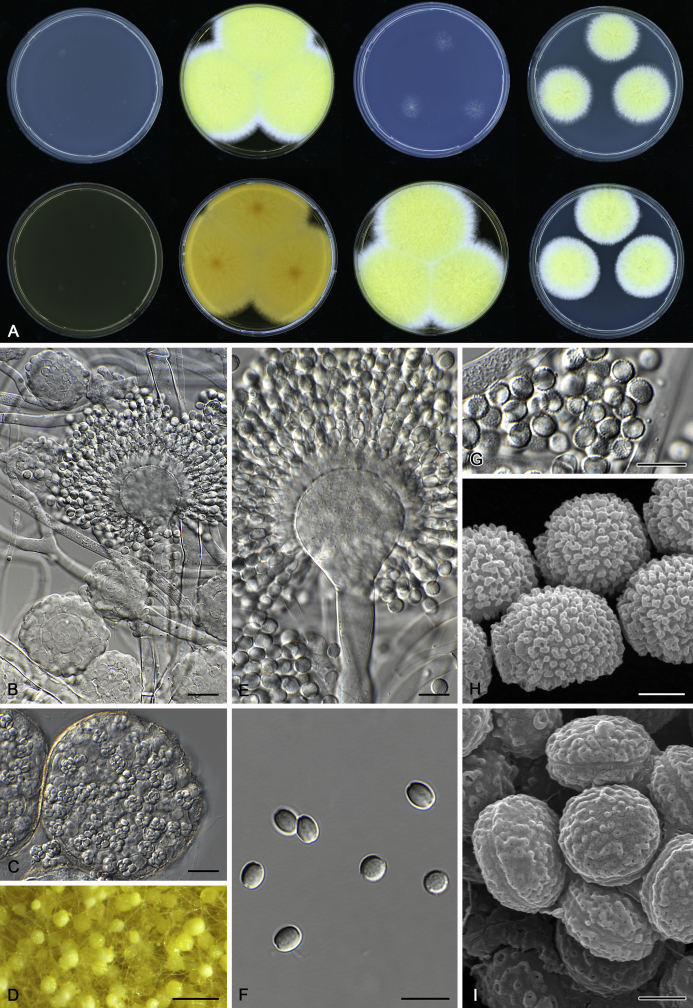
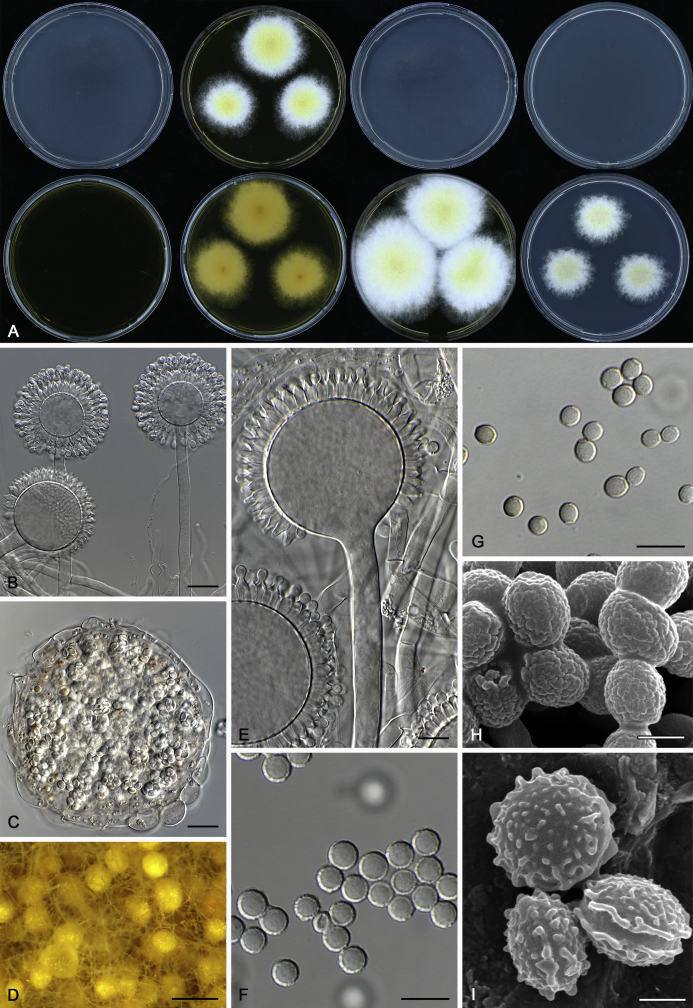
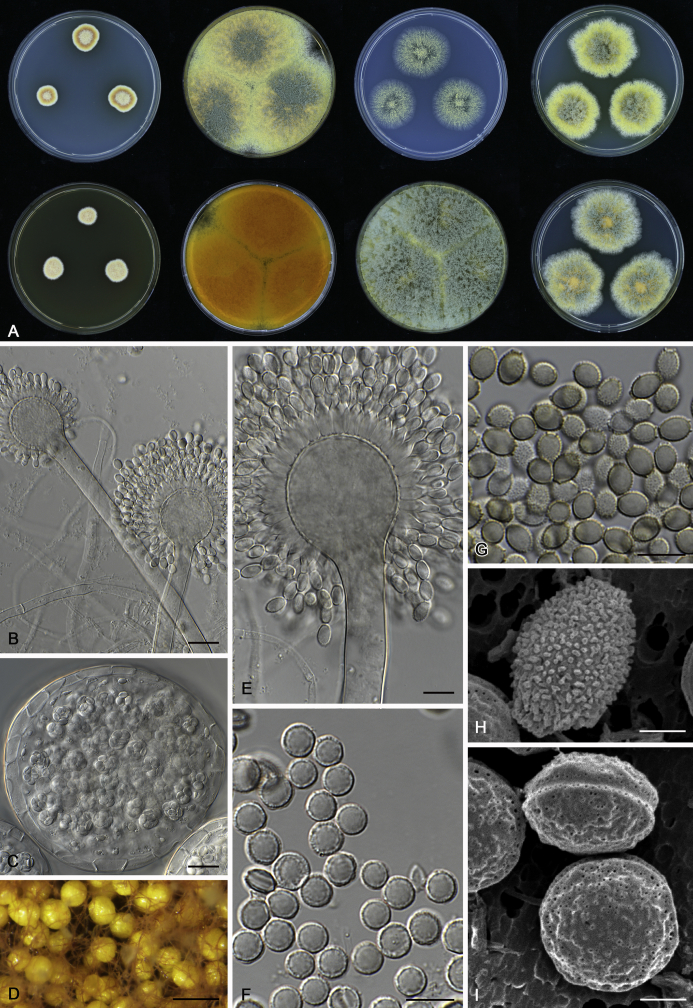
The colony appearance is highly variable within a species. The ratio of asexual and sexual structures can greatly influence the colony appearance (Thom and Raper, 1941, Thom and Raper, 1945, Raper, 1957, Raper and Fennell, 1965). Some strains of A. brunneus, A. sloanii and A. proliferans do not produce sexual structures. On the contrary, anamorphic structures are absent in some strains of A. costiformis and A. cristatus. Hubka et al. (2013a) reported that the anamorph of these species can be induced by decreasing the water activity of the medium and simultaneously raising the incubation temperature. Red-pigmented mycelium was used to distinguish A. ruber from other related species (Raper and Fennell, 1965, Pitt, 1985, Klich, 2002), but can also occur after two weeks in some isolates of A. brunneus, A. cibarius, A. glaucus, A. niveoglaucus, A. proliferans and A. zutongqii (Fig. 13, Fig. 14, Fig. 15, Fig. 16, Fig. 17). Therefore, it cannot be used as a distinguishing character for these species. White conidial heads were used for distinguishing A. niveoglaucus and A. glaucus (Thom and Raper, 1941, Thom and Raper, 1945, Raper and Fennell, 1965), however, green spored A. niveoglaucus strains were reported by Hubka et al. (2013a) and are also confirmed in this study. Other examples include A. montevidensis CBS 410.65 (ex-type of A. heterocaryoticus) and A. ruber CBS 464.65 (ex-type of A. athecius) which produce white or vinaceous buff conidial head, respectively (Fig. 12). Based on these examples, the conidial head colour should not be used as a single distinguishing character either.
Fig. 13.
Growth comparison of Aspergillus sect. Aspergillus species on M40Y for 7 d and 14 d at 25 °C.
Fig. 14.
Growth comparison of Aspergillus sect. Aspergillus species on M40Y for 7 d and 14 d at 25 °C.
Fig. 15.
Growth comparison of Aspergillus sect. Aspergillus species on M40Y for 7 d and 14 d at 25 °C.
Fig. 16.
Growth comparison of Aspergillus sect. Aspergillus species on M40Y for 7 d and 14 d at 25 °C.
Fig. 17.
Growth comparison of Aspergillus sect. Aspergillus species on M40Y for 7 d and 14 d at 25 °C.
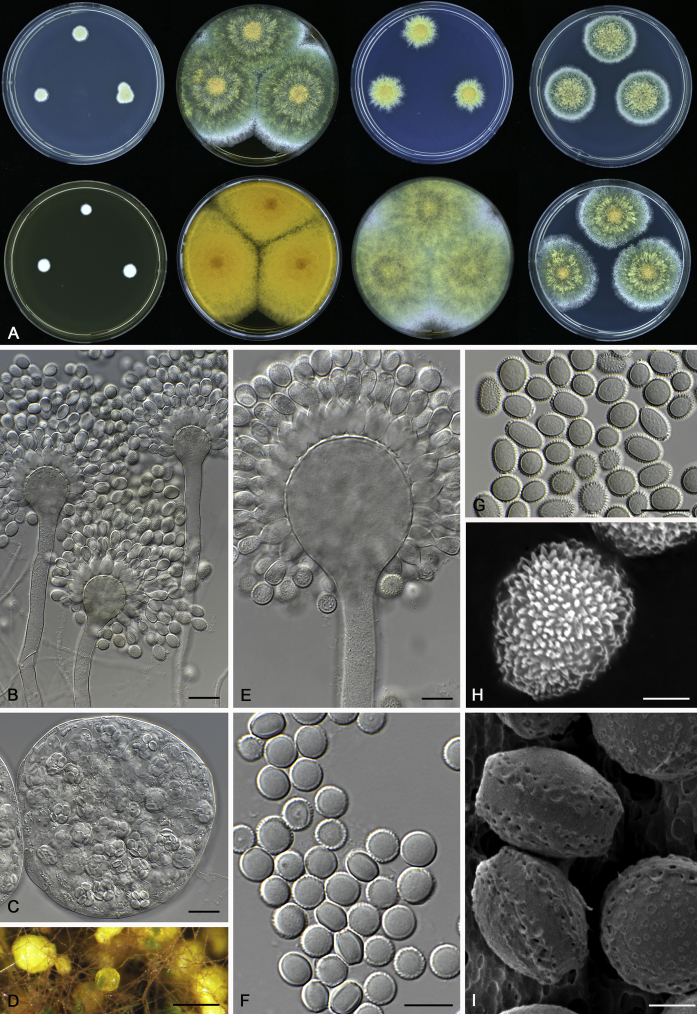
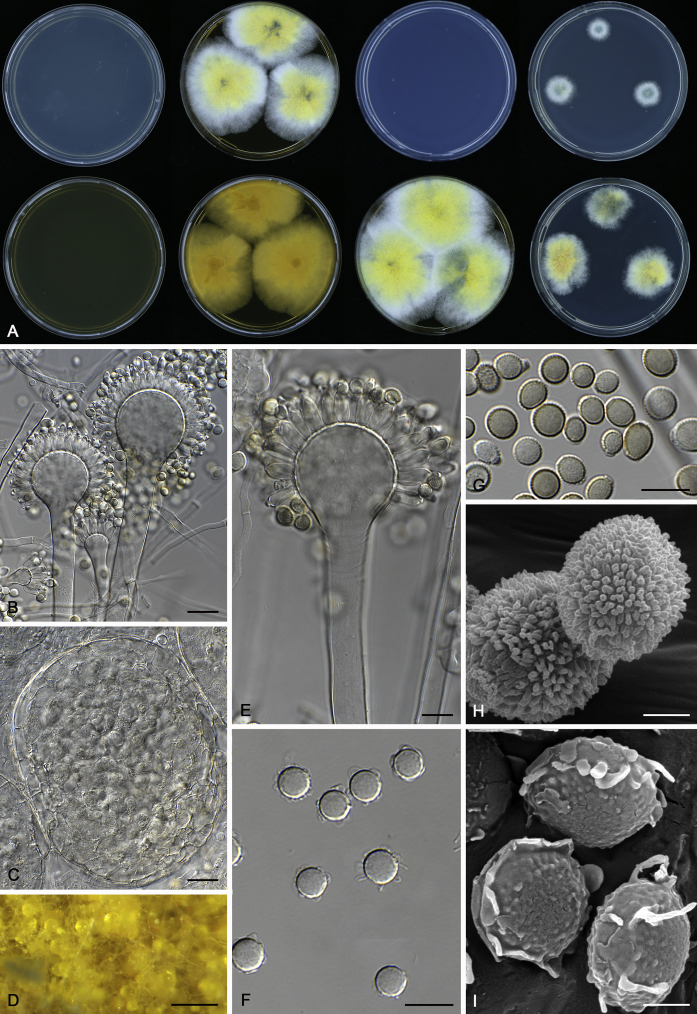
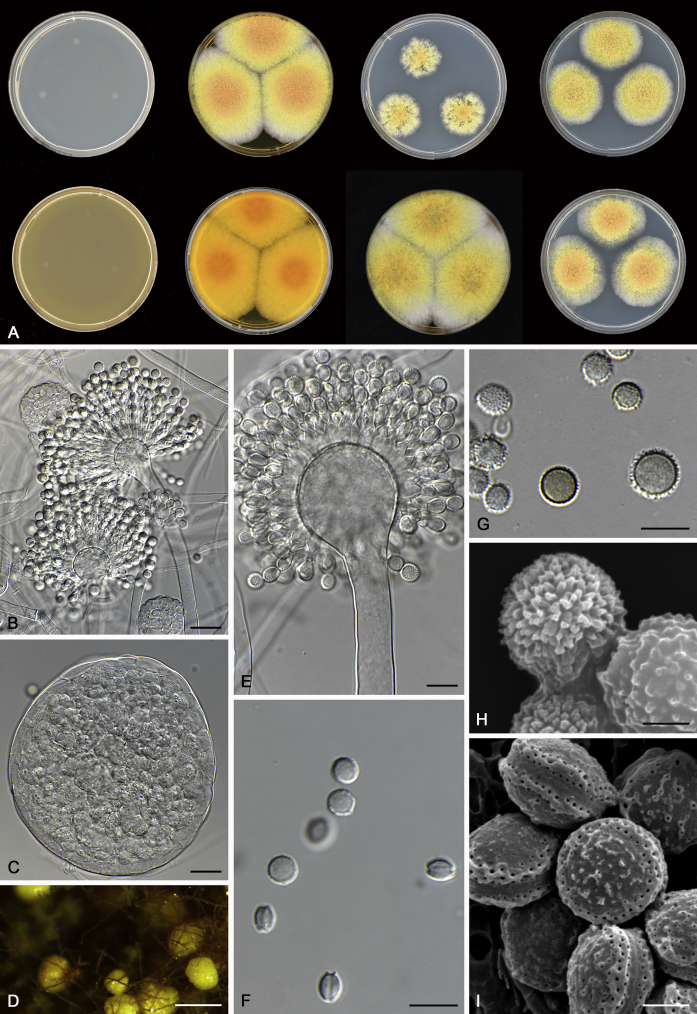
Fig. 12.
Diversity of macromorphology (colonies on M40Y, 25 °C, 7 d) within Aspergillus sect. Aspergillus species. A–E.A. montevidensis. From left to right: CBS 491.65T, CBS 651.74 (ex-type of A. vitis), CBS 410.65, CBS 518.65 (ex-type of A. hollandicus), CBS 111.52. F–J.A. proliferans. From left to right: CBS 121.45T, DTO 322-A2, CCF 4096, CCF 5395, CCF 5392. K–O.A. pseudoglaucus. From left to right: CBS 123.28T, CBS 101747 (ex-type of A. fimicola), CBS 379.75 (ex-type of A. glaber), DTO 147-G3, CGMCC 3.00460. P–T.A. ruber. From left to right: CBS 530.65T, DTO 238-C4, CBS 101748 (ex-type of A. tuberculatus), CBS 104.18, CBS 464.65 (ex-type of A. athecius).
Physiology
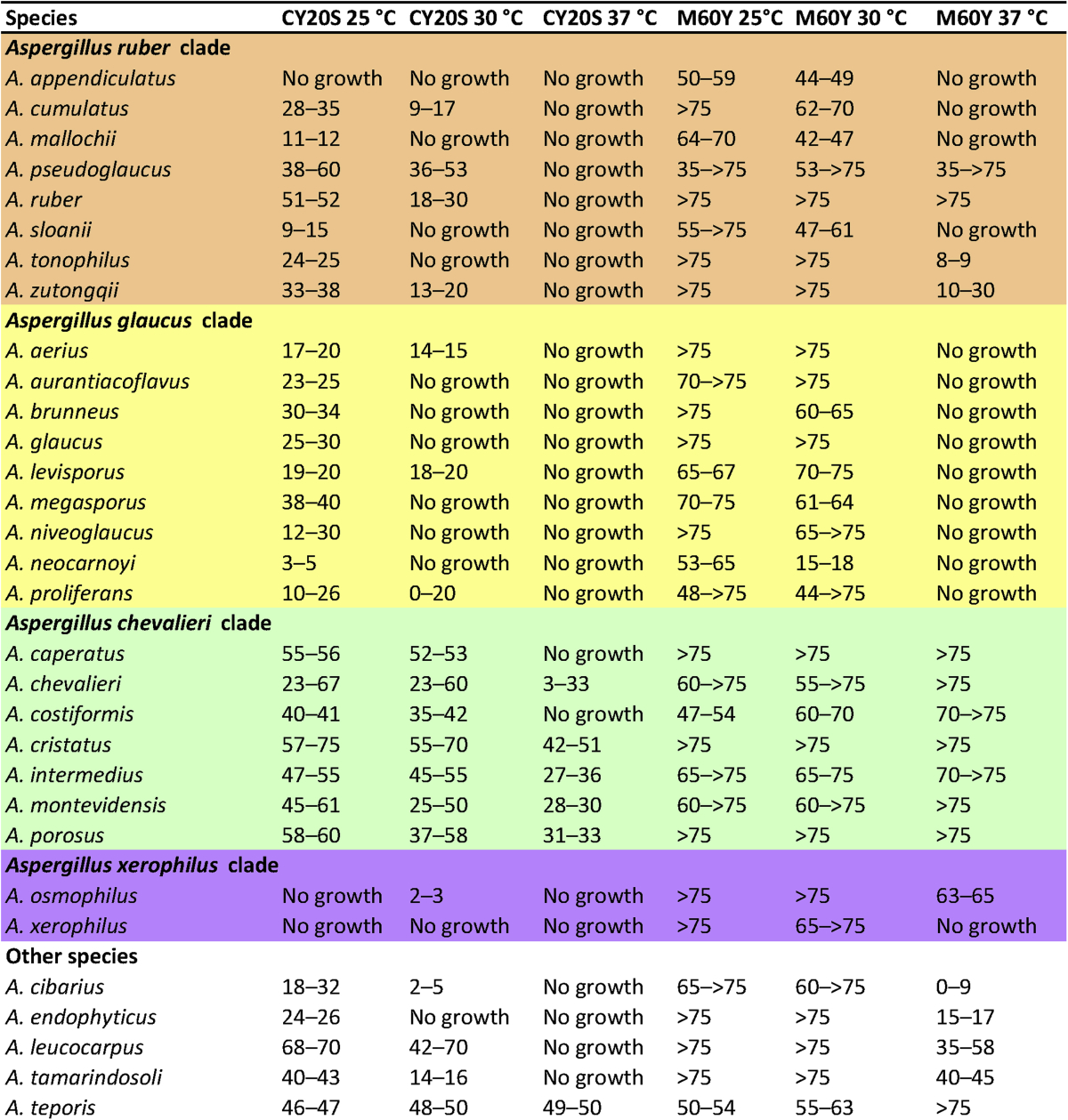
Growth rates at higher temperatures show certain correlation with phylogenetic topologies, most species in the A. chevalieri clade (except A. costiformis and A. caperatus) grow on CY20S at 37 °C, while all species in the A. ruber and A. glaucus clades do not grow under this condition. Growth profiles on M60Y at 37 °C show a similar pattern with CY20S 37 °C. The only difference is that several species from the A. chevalieri and A. ruber clades including A. caperatus, A. costiformis, A. pseudoglaucus, A. ruber, A. tonophilus and A. zutongqii grow on M60Y at 37 °C, but show no growth on CY20S at 37 °C (Table 5). The growth ability on CY20S and M60Y at 37 °C together with the size and surface morphology of ascospores were found to correlate mostly with the phylogenetic species concept in this section (Hubka et al. 2013a). This conclusion is confirmed in this study using a world-wide section Aspergillus strains, and we found the growth ability on CY20S and M60Y at 37 °C a reliable feature for distinguishing morphologically similar species. For example, A. proliferans and A. ruber share similar smooth or slightly rough, furrowed ascospores and tuberculate conidia, among them A. proliferans cannot grow on M60Y at 37 °C, while A. ruber grows well on M60Y at 37 °C. The growth ability on media with high water activity (CYA, MEA) is also useful diagnostic features for certain species. Most species in sect. Aspergillus grow restrictedly on these two media, some species like A. appendiculatus, A. neocarnoyi, A. osmophilus, A. tonophilus and A. xerophilus show more xerophilic abilities compare to others and do not grow on CYA and MEA at all.
Table 5.
Temperature growth profiles (mm) of section Aspergillus species.
Colour codes: yellow = A. glaucus clade, orange = A. ruber clade, green = A. chevalieri clade, purple = A. xerophilus clade.
Micromorphology
Compared to colony appearance, micro-morphological characters within a species are relatively stable and informative (Table 6). The size and ornamentation of ascospores are generally the most informative phenotypic characters for species recognition (Fig. 6, Fig. 7, Fig. 8). Large ascospores (spore bodies average > 6.5 μm) are produced by A. aerius, A. brunneus, A. costiformis, A. glaucus, A. neocarnoyi, A. niveoglaucus, A. osmophilus and A. zutongqii; small ascospores (spore bodies < 5 μm) are produced by A. caperatus, A. intermedius, A. levisporus and A. tamarindosoli, while remaining species produce intermediate ascospores. Convex sides of ascospores can be smooth, verruculose or rugulose, and these ornamentations are generally stable with only minor intraspecific variability. However, in some rare cases, the ascospore morphology differs within a species. For example, most A. ruber strains produce smooth ascospores with minute rough ornamentations along equatorial ridges, but CBS 101748, previously described as A. tuberculatus (Sun & Qi 1994), has tuberculate ascospores (Fig. 8). Variations were also found in A. montevidensis, strain CCF 4248 has similar ascospores with A. tuberculatus, but shows identical sequences, growth parameters and colony phenotype with A. montevidensis (Hubka et al. 2013a), and another atypical strain CCF 4070 has smooth or slightly rough ascospores. It is noteworthy that ascospore ornamentation is related to the stage of development, and fine structures and ornamentation can be overlooked when observed using a light microscope (Blaser, 1975, Kozakiewicz, 1989, Guarro et al., 2012, Hubka et al., 2013a). In addition, some species are morphologically slightly different even when observed under SEM, and therefore careful comparison with experience is needed for morphological identification. Aspergillus parviverruculosus was introduced based on CGMCC 3.04665 producing verruculose ascospores (Kong & Qi 1995), Hubka et al. (2013a) considered it synonymous with A. niveoglaucus based on phylogenetic analysis and they observed appendaged ascospores. We confirmed the appendages in immature ascospores of the ex-type of A. parviverruculosus (CGMCC 3.04665). Filiform appendages were also observed in immature ascospores of A. appendiculatus (Kozakiewicz, 1989, Hubka et al., 2013a) and these appendages can merge with ascospore body and form petaliform crests. Appendaged ascospores are also presented in A. filifer and A. qinqixianii in Aspergillus subgenus Nidulantes; however, these appendages are consistently presented in mature ascospores (Horie et al., 2000, Zalar et al., 2008, Chen et al., 2016).
Table 6.
Most important micromorphological characters for section Aspergillus species (μm).
| Species | Teleomorphic characters |
Anamorphic characters |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Ascomata | Spore bodies | Ornamentation of convex surface | Furrow | Crests | Conidiophores | Vesicles | Phialides | Conidia | |
| Aspergillus aerius | 190–275 | 6.5–8 × 4.5–6 | Smooth, rough along equatorial ridges | Present | Absent | 500–1 000 × 7–15.5 | 26–41 | 7.5–12.5 × 5–8 | Tuberculate, (5–)10–13 × 6–10 |
| A. appendiculatus | 100–225 | 5–7.5 × 4–5.5 | Slightly rough | Absent or showing as a trace | Filiform appendages or petaliform, petals 1–1.5 μm at high parts | 800–2 000 × 7–12(–14.5) | 30–64 | 8–16 × 4.5–7.5 | Tuberculate, 5–10(–12) × 5–7(–8.5) |
| A. aurantiacoflavus | 110–250 | 4–5.5 × 3–5 | Verruculose | Present | Irregular, <0.5 | 250–800 × 7.5–12 | 30–45 | 6–11 × 3.5–6.5 | Tuberculate, 5–9 × 4–7 |
| A. brunneus | 110–240 | 7–10 × 6–8 | Rough along equatorial ridges | Present | Irregular, <0.5 | 700–1 200 × 7–18 | 32–58 | 10–18.5 × 7–12.5 | Tuberculate, 8–15 × 8–13 |
| A. caperatus | 130–220 | 3.5–4.5 × 2.5–4 | Verruculose to rugulose | Pronounced | 0.5–1 | 250–500 × 6.5–9(–12) | 26–45 | 7.5–12 × 4–7.5 | Lobate-reticulate, 3.5–5.5 × 3.5–4.5 |
| A. chevalieri | 100–250 | 3.5–5.5 × 3–4 | Smooth to slightly verruculose | Present | 0.5–1 | 200–1 000 × 6–12 | 23–47 | 5.5–7.5(–10) × 3–5 | Tuberculate to lobate-reticulate, 3–4(–6) × 2.5–3.5(–5) |
| A. cibarius | 100–200 | 4–5.5 × 3–5 | Rough along equatorial ridges | Present | Irregular, <0.5 | 500–700 × 8–14 | 32–58 | 6–11 × 3–5.5 | Tuberculate, 4–7 × 3.5–5.5 |
| A. costiformis | 100–255 | 5.5–7 × 5–6.5 | Rugulose | Pronounced | 0.5 | 500–800 × 7–13 | 20–45(–60) | 6–9.5 × 3–4.5(–5.5) | Microtuberculate, 4–5.5(–6.5) × 3–4.5(–5.5) |
| A. cristatus | 100–200 | 4.5–6 × 4–6 | Verruculose to rugulose | Present | 1.2–1.5 | 300–500 × (6–)8–12 | (26–)35–51 | 5.5–9 × 3.5–6 | Tuberculate, 4–6.5 × 3.5–5 |
| A. cumulatus | 100–200 | 4–6 × 3.5–5 | Slightly rough | Pronounced | Irregular, <0.5 | 500–1 300 × 7–15 | 32–57 | 7–12 × 4.5–7.5 | Tuberculate, 5–8 × 4–7.5 |
| A. endophyticus | 120–200 | 4–5.5 × 3–4.5 | Verruculose to rugulose | Pronounced | 0.5–1 | 350–800 × 9.5–14 | 32–52 | 6–10 × 3.5–5.5 | Tuberculate to lobate-reticulate, 5.5–8 × 4.5–6 |
| A. glaucus | 120–250 | 5.5–7.5 × 3.5–6 | Smooth, minute rough along equatorial ridges | Pronounced | Irregular, 0.5–1 | 150–500 × 10–21(–30) | 30–60 | (8–)12–20 × (4–)5–8.5 | Tuberculate, 6–12.5 × 5.5–9 |
| A. intermedius | 100–250 | 3.5–5 × 3–4.5 | Verruculose to rugulose | Present | 0.5 | 250–600 × 7.5–13 | (26–)40–60 | 5.5–7.5(–9) × 3–5.5 | Microtuberculate, 3–4(–6) × 3–4.5 |
| A. leucocarpus | 80–140 | 4.5–5.5 × 3.5–5 | Verruculose | Present | 0.8–1.5 | 800–1 400 × 7.5–12 | 35–60 | 8–11.5 × 3.5–6.5 | Tuberculate, 5.5–9 × 5–8 |
| A. levisporus | 70–130 | 3–4.5 × 2.5–4 | Smooth | Present | Absent | 400–600 × 10–14 | 30–44 | 6–8.5 × 3.5–6 | Tuberculate to lobate-reticulate, 3.5–4.5 × 2.5–4 |
| A. mallochii | 130–220 | 4–6 × 3–5 | Smooth, minute rough along equatorial ridges | Absent or showing as a trace | Petaliform, 1–2 at high parts | 600–1 500 × 6–9.5(–12) | 27–43 | 6.5–9 × 3–5 | Tuberculate, 4.5–7 × 4–5.5 |
| A. megasporus | 110–300 | 4–6.5 × 3.5–5.5 | Smooth, rough along equatorial ridges | Present | Absent or indefinite | 1 000–1 500 × 6.5–12(–21.5) | 30–54 | 7.5–14 × 4–7.5 | Tuberculate, 7–14 × 5–8.5 |
| A. montevidensis | 80–250 | 4–6 × 3–4.5 | Generally rugulose, smooth or slightly rough in atypical strain CCF 4070, tuberculate in atypical strain CCF 4248 | Pronounced | 0.5 | 250–500 × 6–13.5 | 25–35(–50) | 5–8.5(–11) × 3–6 | Lobate-reticulate, 4–6.5 × 3.5–5 |
| A. neocarnoyi | 120–230 | 6.5–9 × 4.5–7 | Verruculose to rugulose | Present | Absent or indefinite | 1 000–2 000 × (9–)12–23 | (32–)50–92 | 12–21 × 6–9 | Tuberculate, 8–15.5 × 6–10 |
| A. niveoglaucus | 90–240 | (4.5–)5.5–7.5 × (3–)5–6 | Rough along equatorial ridges or verruculose to rugulose | Present | Irregular, <0.5 | 1 000–1 500 × (7.5–)10–23 | (31–)55–85 | 8–14(–20) × 4–7(–11) | Tuberculate, (6–)8–13.5 × 4–9 |
| A. osmophilus | 100–350 | 7–9 × 6–7.5 | Verruculose | Pronounced | 0.5 | 300–1 000 × 7.5–12 | 28–46 | 9–12 × 4.5–7 | Microtuberculate to tuberculate, 6–8.5 × 5.5–7.5 |
| A. porosus | 80–230 | 3.5–5.5 × 3–4.5 | Rugulose, pitted | Pronounced | 0.5 | 250–600 × 5–12.5 | 24–58 | 5–10 × 2.5–5 | Lobate-reticulate, 3.5–5.5 × 2.5–4.5 |
| A. proliferans | 100–240 | 4–6 × 3–5 | Smooth or slightly verruculose or rough along equatorial ridges | Present or pronounced | Absent | 250–1 000 × 8–16.5 | 20–50 | 6–12 × 3–5.5 | Tuberculate, 5–7.5(–10) × 4–6(–7) |
| A. pseudoglaucus | 75–200 | 4–6.5 × 3–4.5 | Smooth or slightly rough | Absent or showing as a trace | Absent | 500–1 000 × (7–)11–22 | (26–)37–65 | 6–11 × 4–6.5 | Tuberculate; microtuberculate in atypical strain CBS 379.75, (3.5–)6–9 × (3–)5.5–7.5 |
| A. ruber | 50–175 | 4–6 × 3.5–5 | Generally smooth or minute rough along equatorial ridges, tuberculate in atypical strain CBS 101748 | Present or pronounced | Absent | 500–750 × 7–13.5 | 25–48 | 7–9(–12) × 3.5–6 | Tuberculate, (4.5–)7–9(–12) × 4–6(–8) |
| A. sloanii | 60–205 | 4–6 × 3–4.5 | Smooth, minute rough along equatorial ridges | Present | Absent | 160–900 × 7.5–16 | (10–)34–53 | (7.5–)9–13.5(–18) × (5–)7–9.5 | Tuberculate, 5.5–9.5 × 5.5–9 |
| A. tamarindosoli | 130–240 | 3.5–5 × 3–4 | Verruculose | Present | Irregular, 0.5–1.5 | 700–1 000 × 10–15 | 40–72 | 6.5–12 × 4–5.5 | Lobate-reticulate, 4–7 × 3–4.5 |
| A. teporis | 120–180 | 5–6.5 × 4–5.5 | Slightly verruculose | Pronounced | 0.5 | 800–1 200 × 8–19 | 33–53 | 7–12 × 3.5–5 | Lobate-reticulate, 3.5–6 × 3–4.5 |
| A. tonophilus | 100–235 | 4–6 × 3–4.5 | Verruculose | Present | Absent | 120–500 × 7–12.5 | 25–44 | 6–11 × 3–5 | Tuberculate to lobate-reticulate, 5–7.5 × 3.5–6 |
| A. xerophilus | 165–330 | 4.5–6.5 × 3.5–5 | Verruculose | Present | Irregular, <0.5 | 50–200 × 6.5–9.5(–12) | 40–66 | 6–9 × 3.5–6 | Microtuberculate, 3.5–5.5 × 3–4.5 |
| A. zutongqii | 110–220 | 6–7.5 × 4.5–6 | Verruculose | Pronounced | Absent | 150–500 × 7.5–13 | 25–40 | 8–12 × 4–6.5 | Tuberculate, 5.5–10 × 4–7 |
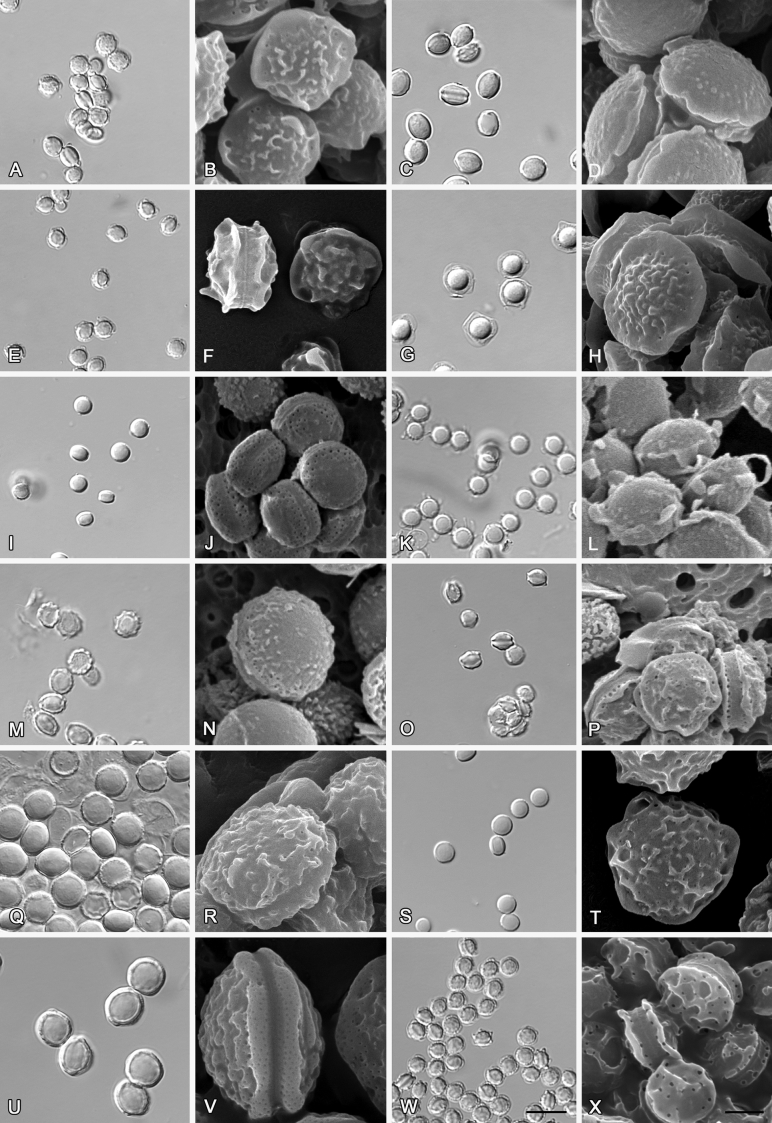
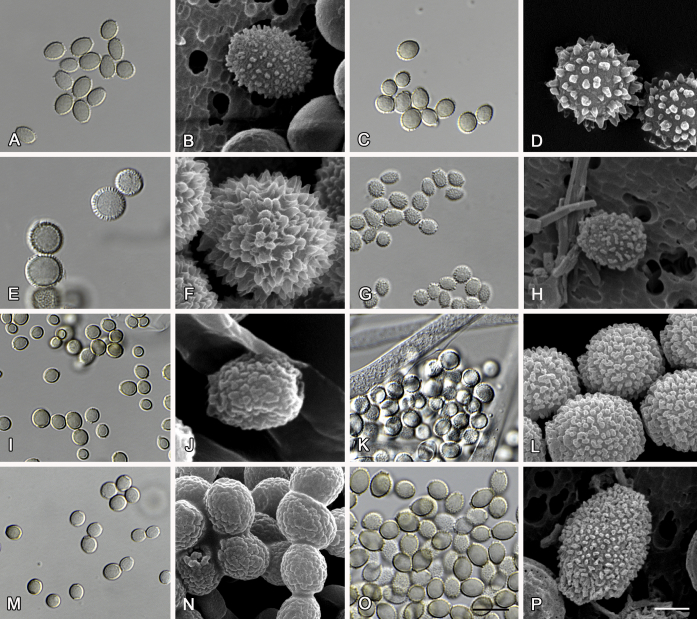
Fig. 6.
Formation of ascomata and range of ascospore phenotypes. A–D. Initials and ascomata. E, F.Aspergillus aerius CBS 141771T. G, H.A. appendiculatus CBS 374.75T. I, J.A. aurantiacoflavus CBS 141930T. K, L.A. brunneus CBS 112.26T. M, N.A. caperatus CBS 141774T. O, P.A. chevalieri CBS 522.65T. Q, R.A. cibarius KACC 46346T. S, T.A. costiformis CBS 101749T. U, V.A. cristatus CBS 123.53T. W, X.A. cumulatus KACC 47316T. Scale bars: D = 20 μm, applies to A–C; W = 10 μm, applies to E, G, I, K, M, O, Q, S, U; X = 2 μm, applies to F, H, J, L, N, P, R, T, V.
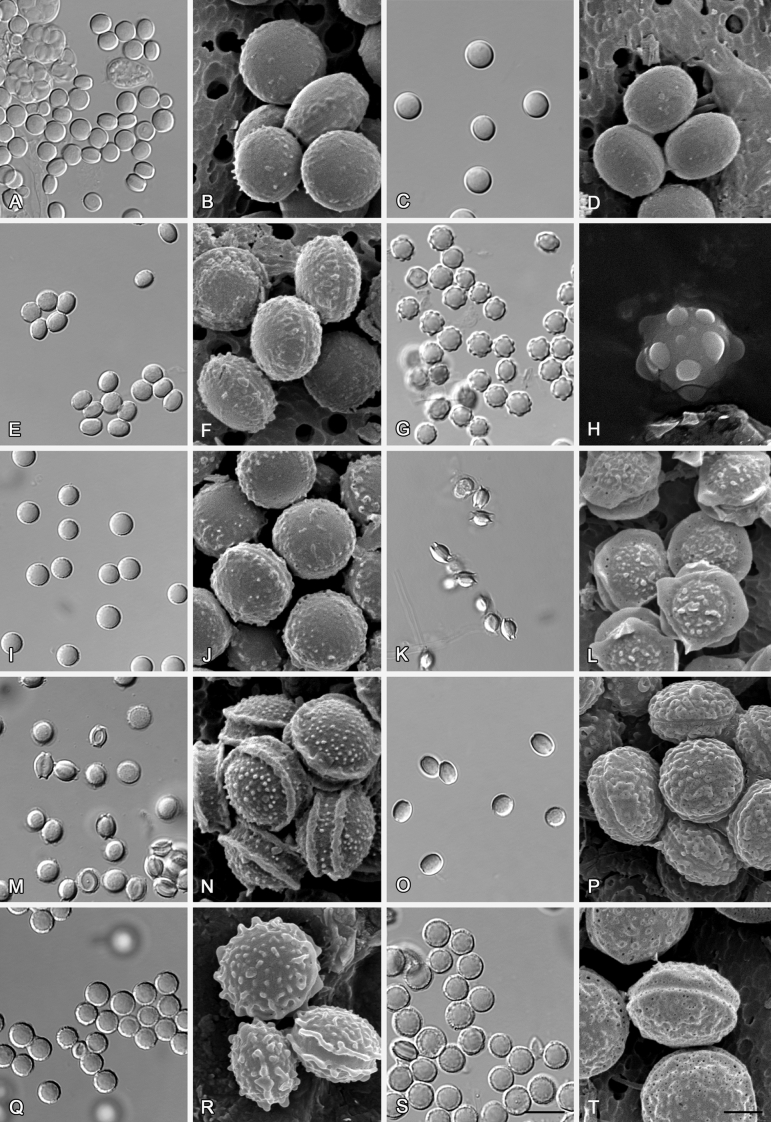
Fig. 7.
Range of ascospore phenotypes. A, B.Aspergillus endophyticus CBS 141766T. C, D.A. glaucus CBS 516.65T. E, F.A. intermedius CBS 523.65T. G, H.A. leucocarpus CBS 353.68T. I, J.A. levisporus CBS 141767T. K, L.A. mallochii CBS 141928T. M, N.A. megasporus CBS 141929T. O, P.A. montevidensis CBS 491.65T. Q, R.A. neocarnoyi CBS 471.65T. S, T.A. niveoglaucus CBS 114.27T. U, V.A. osmophilus CBS 134258T. W, X.A. porosus CBS 141770T. Scale bars: W = 10 μm, applies to A, C, E, G, I, K, M, O, Q, S, U; X = 2 μm, applies to B, D, F, H, J, L, N, P, R, T, V.
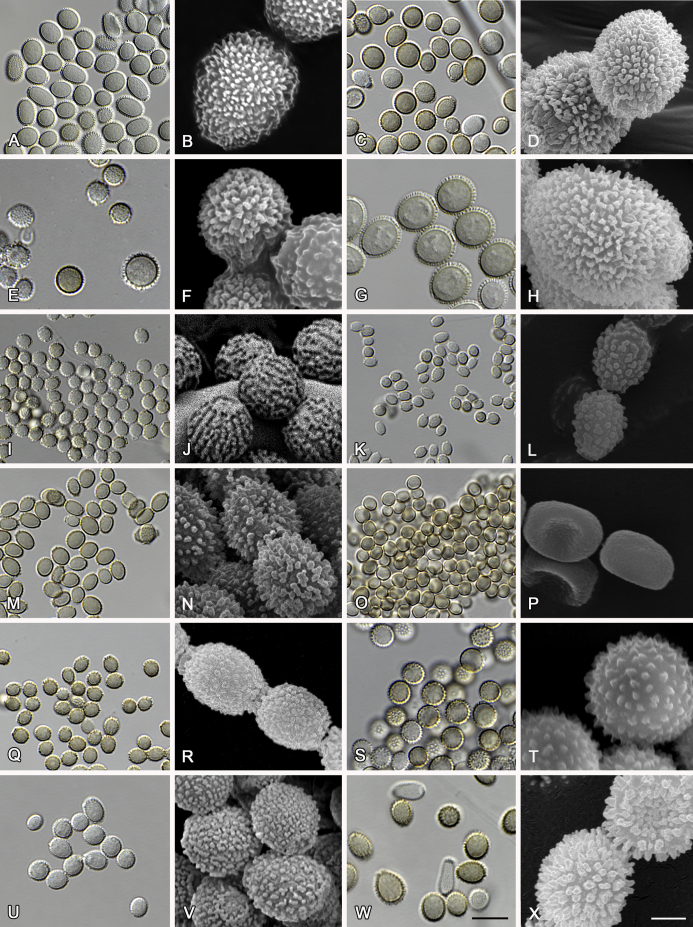
Fig. 8.
Range of ascospore phenotypes. A, B.Aspergillus proliferans DTO 322-A2. C, D.A. pseudoglaucus CBS 101747 (ex-type of A. fimicola). E, F.A. ruber CBS 530.65T. G, H.A. ruber CBS 101748 (ex-type of A. tuberculatus). I, J.A. sloanii CBS 138177T. K, L.A. tamarindosoli CBS 141775T. M, N.A. teporis CBS 141768T. O, P.A. tonophilus KACC 47150. Q, R.A. xerophilus CBS 938.73T. S, T.A. zutongqii CBS 141773T. Scale bars: S = 10 μm, applies to A, C, E, G, I, K, M, O, Q; T = 2 μm, applies to B, D, F, H, J, L, N, P, R.
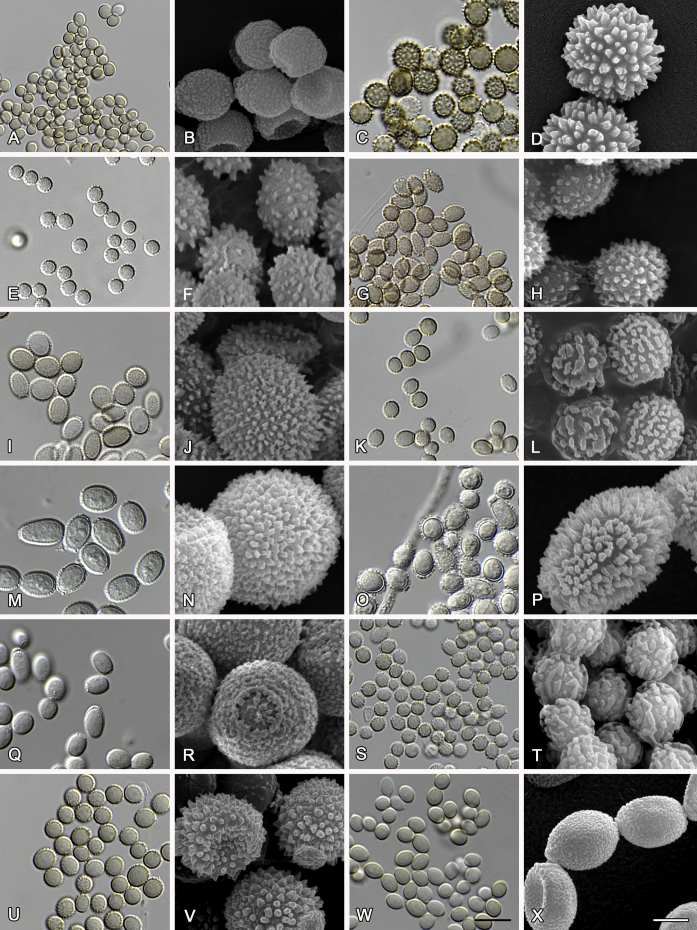
The diameter and shape of conidia are highly variable within species and generally not useful for species differentiation. However, conidial ornamentation is useful for differentiating phylogenetically related species or species with similar ascospore morphology (Fig. 9, Fig. 10, Fig. 11). For example, A. intermedius is phylogenetically related to A. montevidensis. Both produce verruculose ascospores with 0.5 μm crests, however, the microtuberculate conidia of A. intermedius can easily distinguish it from A. montevidensis. Most species produce consistent conidial ornamentations, except in A. pseudoglaucus where most strains produce tuberculate conidia, but CBS 379.75, previously described as A. glaber (Blaser 1975), produces microtuberculate conidia. Kozakiewicz (1989) assigned the conidial ornamentation into four categories, ranging from microtuberculate, aculeate, tuberculate to lobate-reticulate. Based on our observations, aculeate and tuberculate ornamentations may occur in same species, and can be affected by the fixation methods or age of conidia. We therefore, combined these two types of ornamentation within the tuberculate category. The three categories of conidial ornamentation described here include microtuberculate, tuberculate or lobate-reticulate.
Fig. 9.
Range of conidia phenotypes. A, B.Aspergillus aerius CBS 141771T. C, D.A. appendiculatus CBS 374.75T. E, F.A. aurantiacoflavus CBS 141930T. G, H.A. brunneus CBS 112.26T. I, J.A. caperatus CBS 141774T. K, L.A. chevalieri CBS 522.65T. M, N.A. cibarius KACC 46346T. O, P.A. costiformis CBS 101749T. Q, R.A. cristatus CBS 123.53T. S, T.A. cumulatus KACC 47316T. U, V.A. endophyticus CBS 141766T. W, X.A. glaucus CBS 516.65T. Scale bars: W = 10 μm, applies to A, C, E, G, I, K, M, O, Q, S, U; X = 2 μm, applies to B, D, F, H, J, L, N, P, R, T, V.
Fig. 10.
Range of conidia phenotypes. A, B.Aspergillus intermedius CBS 523.65T. C, D.A. leucocarpus CBS 353.68T. E, F.A. levisporus CBS 141767T. G, H.A. mallochii CBS 141928T. I, J.A. megasporus CBS 141929T. K, L.A. montevidensis CBS 491.65T. M, N.A. neocarnoyi CBS 471.65T. O, P.A. niveoglaucus CBS 114.27T. Q, R.A. osmophilus CBS 134258T. S, T.A. porosus CBS 141770T. U, V.A. pseudoglaucus CBS 101747 (ex-type of A. fimicola). W, X.A. pseudoglaucus CBS 379.75 (ex-type of A. glaber). Scale bars: W = 10 μm, applies to A, C, E, G, I, K, M, O, Q, S, U; X = 2 μm, applies to B, D, F, H, J, L, N, P, R, T, V.
Fig. 11.
Range of conidia phenotypes. A, B.Aspergillus proliferans DTO 322-A2. C, D.A. ruber CBS 530.65T. E, F.A. sloanii CBS 138177T. G, H.A. tamarindosoli CBS 141775T. I, J.A. teporis CBS 141768T. K, L.A. tonophilus KACC 47150. M, N.A. xerophilus CBS 938.73T. O, P.A. zutongqii CBS 141773T. Scale bars: O = 10 μm, applies to A, C, E, G, I, K, M; P = 2 μm, applies to B, D, F, H, J, L, N.
Extrolites
Species in sect. Aspergillus produce some main biosynthetic families of secondary metabolites. All species of sect. Aspergillus produce echinulins and derived isoechinulins and neoechinulins. Aspergillus sloanii is the only species that does not convert echinulins to isoechinulins and neoechinulins (Table 7, Table 8). Furthermore, the echinulin related molecules arestrictin A & B and cristatin A are produced by A. restrictus and A. penicillioides in sect. Restricti (Itabashi et al. 2006). Certain polyketides are also commonly detected in sect. Aspergillus, including octaketide anthraquinones, such as emodin and physcion. Other compounds commonly detected include anthraquinones, and the related asperflavin. They are absent from A. montevidensis, which explain the bright yellow colour of its ascomata. Species that have orange-red or red mycelium covering the ascomata, such as A. ruber, produce additional red anthraquinones, including erythroglaucin and catenarin. Emodin and physcion and their bisanthrons are found to be common. These compounds were detected in the closely related species in sect. Cremei, for example in A. wentii (Wells et al., 1975, Assante et al., 1980, Rabie et al., 1986). Also, A. stromatioides in sect. Cremei produces emodin and ω-hydroxyemodin, which is shared with sect. Aspergillus (González-Andrade et al. 2013). Another octaketide, sulochrin is only recovered from A. xerophilus. Sulochrin and similar compounds, i.e., 3-O-methylsulochrin and 3-O-demethylsulochrin have also been recovered from A. wentii and A. europaeus in sect. Cremei (Rabie et al., 1986, Hubka et al., 2016). The octaketide asperentin was reported first from A. flavus (Grove 1972a), but the fungus was misidentified and was actually A. pseudoglaucus. Aspergillus brunneus and A. neocarnoyi can also produce asperentins.
Table 7.
Extrolites reported from Aspergillus section Aspergillus.1
| Biosynthetic family | Compounds | References | Producers |
|---|---|---|---|
| Cyclic dipeptides with a dimethylallyl group | LL-S-490β = N-a-acetylaszonalenine, rugulosuvine | Ellestad et al., 1973, Micheluz et al., 2016 | A. glaucus, A. pseudoglaucus |
| Deoxybrevianamides2 | Deoxybrevianamide E | Micheluz et al. 2016 | A. glaucus (& A penicillioides) |
| Stachybotryamides2 | Stachybotryamide | Micheluz et al. 2016 | A. glaucus |
| Tryprostatins2 | Tryprostatin B | Micheluz et al. 2016 | A. glaucus |
| Tenellins2 | Tenellin | Micheluz et al. 2016 | A. glaucus |
| Echinulins | Echinulin, dehydroechinulin, didehydroechinulin, preechinulin, L-alanyl-L-tryptophan anhydride, (L-valyl-L-tryptophan anhydride, cryptoechinulin G, neoechinulin, neoechinulin A, neoechinulin B (= E-10), neoechinulin C (= cryotoechinulin A = E-8), neoechinulin D, neoechinulin E = cryptoechinulin C, dihydroneoechinulin B, isoechinulin A, isoechinulin B, isoechinulin C, dihydroxyisoechinulin A, rubrumazine A, rubrumazine B, rubrumazine C, tardioxopiperazine A, tardioxopiperazine B, dehydrovariecolorin L, variecolorin A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, golmaneone, 12-demethyl-12-oxo-eurotechinulin A, B, alkaloid E-7, cristatumin A, cristatumin B, cristatumin C,3 cristatumin D, cristatumin E,3 critatumin F, eurocristatine3 | Quilico and Panazzi, 1943, Quilico and Cardini, 1950, Barbetta et al., 1969, Nakashima and Slater, 1971, Allen, 1972, Dossena et al., 1974, Cardillo et al., 1974, Cardillo et al., 1975, Marchelli et al., 1975, Nagasawa et al., 1975, Nagasawa et al., 1976, Hamasaki et al., 1976a, Hamasaki et al., 1976b, Stipanovic and Schroeder, 1976, Stipanovics et al., 1976, Inoue et al., 1977a, Marchelli et al., 1977, Gatti et al., 1978, Podojil et al., 1979, Fujimoto et al., 1999 (fungus was A. pseudoglaucus, misidentified as Microascus tardifaciens), Li et al., 2004a, Li et al., 2004b, Smetanina et al., 2007, Wang et al., 2007a, Wang et al., 2007b, Wang et al., 2007c, Li et al., 2008a, Li et al., 2008b, Slack et al., 2009, Almeida et al., 2010, Zhou et al., 2010 (misidentified as a Penicillium griseofulvum); Du et al., 2012, Gao et al., 2011, Gao et al., 2012a, Gomes et al., 2012, Yan et al., 2012, Gao et al., 2013 (misidentified as A. effusus), Wu et al., 2013, Miyake et al., 2014, Zou et al., 2014, Meng et al., 2015, Micheluz et al., 2016, Visagie et al., 2017 | A. brunneus, A. chevalieri, A. cristatus, A. glaucus, A. mallochii, A. megasporus, A. montevidensis, A. proliferans, A. pseudoglaucus, A. ruber (Arestricticins A, B, cristatin A and asperglaucide were reported from A. restrictus and A. penicillioides (Itabashi et al. 2006), indicating a strong relationship between species in sections Aspergillus and Restricti.) |
| Quinolone | Quinolactacin A1, Quinolactacin A2, Quinolactacin B | Visagie et al. 2017 | A. megasporus |
| Chimeric echinulins and auroglaucins | Cryptoechinulin B (= aurechinulin), (+) and (−)-cryptoechinulin D, 7-O-methylvariecolortide A, variecolortide A, (+) & (−) variecolortide B, (+) & (−) variecolortide C, (+) & (−) 7-isopentenylcryptoechinuline D, dihydrocryptoechinulin D, effusin A | Gatti et al., 1976, Inoue et al., 1977b, Li et al., 2010, Wang et al., 2007b (misidentified as A. variecolor); Kuttruff et al., 2011, Yan et al., 2012, Gao et al., 2012b (misidentified as A. effusus), Gao et al. 2013 (misidentified as A. effusus), Chen et al. 2014 | Aspergillus montevidensis, A. ruber |
| Anthraquinones | Catenarin, emodic acid, emodin (= parietin), erythroglaucin, fallacinol, physcion, questin (= emodin 8-O-methylether), questinol, rubrocristin, variecolorquinone A, (2S)-2,3-dihydroxypropyl-1,6,8-trihydroxy-3-methyl-9,10-dioxoanthracene-2-carboxylate, 3-O-(α-D-ribofuranosyl)questinol, 3-O-(α-D-ribofuranosyl)questin, rubrocristin, viocristin, isoviocristin, hydroxyviocristin, eurorubrin, asperinine A, B, ω-hydroxyemodin-5-merthyether, ω-hydroxyrubrocristin | Anslow and Raistrick, 1940, Bachmann et al., 1979, Bachmann et al., 1982, Anke et al., 1980a, Anke et al., 1980b, Fujimoto et al., 1999, Engstrom et al., 1982, Laatsch and Anke, 1982, Arai et al., 1989, Wang et al., 2006 (as Chaetomium globosum), Smetanina et al., 2007, Wang et al., 2007c (fungus misidentified as A. variecolor), Du et al., 2008, Wang et al., 2008, Li et al., 2009, Gomes et al., 2012, Almeida et al., 2010, Yan et al., 2012, Du et al., 2014, Micheluz et al., 2016, Visagie et al., 2017 | A. brunneus, A. chevalieri, A. cristatus, A. glaucus, A. intermedius, A. leucocarpus, A. mallochii, A. megasporus, A. neocarnoyi, A. niveoglaucus, A. pseudoglaucus, A. ruber, A. tonophilus (Citreorosein was reported from A. penicillioides by Micheluz et al. 2016) |
| Asperflavins | Anhydroasperflavin, asperflavin, asperflavin ribofuranoside, isoasperflavin | Grove 1972a (misidentified as A. flavus), Anke et al., 1978, Fujimoto et al., 1999, Li et al., 2006 (misidentified as Microsporum), Smetanina et al., 2007, Du et al., 2008, Du et al., 2014 | A. glaucus, A. pseudoglaucus, A. megasporus |
| Isotorachrysones | Isotorachrysone, isotarachrysone 6-O-α-D-ribofuranoside, 8-methoxy-3-methyl-1-naphthalenol-6-O-α-D-ribofuranoside, 8-methoxy-1-naphthalenol-6-O-α-D-ribofuranoside, (+)-variecolorquinone A, aspergiodiquinone | Wang et al. 2007a (misidentified as A. variecolor), Du et al., 2008, Sun et al., 2013 | A. glaucus |
| Aspergiolides | Aspergiolide A, B, C, D | Du et al., 2007, Du et al., 2008, Du et al., 2011, Sun et al., 2009, Sun et al., 2013, Tao et al., 2009 | A. glaucus |
| Eurotionones | Eurotinone, variecolorquinone B, 2-methyleurotinone, 9-dehydroxyeurotinone, 2-O-methyl-9-dehydroxyeurotinone, 2-O-methyl-4-O-(α-D-ribofuranosyl)-9-dehydroxyeurotinone, all related to anthraquinones, (+) & (−) europhenol A | Wang et al., 2007c, Li et al., 2009, Yan et al., 2012, Miyake et al., 2014, Meng et al., 2016 | A. ruber |
| Bianthrons | (trans) & (cis)-emodin-physcion bianthrone, physcionanthrone (= physciondianthranol), physcion bianthrone (= physcion-dianthrone = physcion anthrone dimer), physcion-anthrone A (= physcion-9-anthrone), physcion anthrone B | Ashley et al., 1939, Bachmann et al., 1979, Anke et al., 1980a, Anke et al., 1980b | A. cristatus, A. chevalieri |
| Asperentins | Asperentin (=cladosporin), cladosporin 8-O-methylether (= asperentin 8-O-methyl ether), aspyran, asperentin-6-O-methyl ether, 5′-hydroxy-asperentin-8-methyl ether, 5′-hydroxyasperentin, 4′-hydroxyasperentin, (isocladosporin), 6-O-α-D-ribosylasperentin, 6-O-α-D-ribosyl-8-O-methylasperentin, 5-hydroxyl-6-O-methylasperentin | Grove, 1972b, Grove, 1973, Cattel et al., 1973 (misidentified as A. flavus), Podojil et al., 1979, Fujimoto et al., 1999, Wang et al., 2006 (as Chaetomium globosum), Slack et al., 2009, Kimura et al., 2012, Wang et al., 2013 (as Cladosporium cladosporioides), Kozlovsky et al., 2014, Tang et al., 2014, Greco et al., 2015, Cochrane et al., 2016 (from Cladosporium cladosporioides) | A. chevalieri, A. montevidensis, A. proliferans, A. pseudoglaucus, A. ruber |
| Mycophenolic acids | 5,7-dihydroxy-4-methylphthalide, 6-farnesyl-5,7-dihydroxy-4-methylphthalide , mycophenolic acid | Grove, 1972a, Grove, 1972b (misidentified as A. flavus), Burkin and Kononenko, 2010, Gao et al., 2011, Gao et al., 2012a, Gao et al., 2012b, Séguin et al., 2014 | A. pseudoglaucus, A. ruber (traces of mycophenolic acid), A. montevidensis (traces of mycophenolic acid), A. chevalieri (traces of mycophenolic acid) |
| Pseurotins2 | Pseurotin A & D | Micheluz et al. 2016 | A. glaucus |
| Orsellinic acid derivatives | Cristatumside A | Du et al. 2014 | A. cristatus |
| Kotanins | Desmethylkotanin, kotanin | Büchi et al. 1971 | A. glaucus |
| Auroglaucins | Auroglaucin, flavoglaucin, dihydroauroglaucin, isodihydroauroglaucin, isotetrahydroauroglaucin (= dihydroflavoglaucin), chaetopyranin, 2-(2′,3-epoxy-1′,3-heptadienyl)-6-hydroxy-5-(3-methyl-2-butenyl)benzaldehyde, tetrahydroauroglaucin, (E)-2-(hept-1-enyl)-3-(hydroxymethyl)-5-(3-methylbut-2-enyl)benzene-1,4-diol, (E)-4-(hept-1-enyl)-7-(3-methylbut-2-enyl)-2,3-dihydrobenzofuran-2,5-diol, eurotirumin, 2-(2′,3-epoxy-1′-heptenyl)-6-hydroxy-5-(3″-methyl-2″-butenyl)benaldehyde, (E)-6-hydroxy-7-(3-methyl-2-butenyl)-2-(3-oxobut-1-enyl)chroman-5-carbaldehyde, 2-(1′,5′-heptadienyl)3,6-dihydroxy-5-(3″-methyl-2″butenyl)benzaldehyde, aspergentisyl A, B, aspergin | Gould and Raistrick, 1934, Ashley et al., 1939, Quilico et al., 1949, Birch, 1958, Inoue et al., 1977c, Hamasaki et al., 1980, Hamasaki et al., 1981, Ishikawa et al., 1984, Ishikawa et al., 1985, Li et al., 2006, Wang et al., 2006 (as Chaetomium globosum), Li et al., 2008a, Li et al., 2008b, Miyake et al., 2009, Slack et al., 2009, Almeida et al., 2010, Miyake et al., 2010a, Miyake et al., 2010b, Gao et al., 2011, Gao et al., 2012a, Gao et al., 2012b, Gao et al., 2013 (misidentified as A. effusus), Sun et al., 2013, Wu et al., 2013, Miyake et al., 2014, Visagie et al., 2017 | A. brunneus, A. glaucus, A. glaucus? (as Microsporum sp.), A. chevalieri, A. cristatus, A glaucus, A. montevidensis, A. mallochii, A. pseudoglaucus, A. ruber |
| Heveadrides | Epiheveadride, heveadride | Slack et al. 2009 | A. glaucus, A. montevidensis, A. ruber |
| Chaetoviridins | Chaetoviridin A | Micheluz et al. 2016 | A. glaucus |
| Eurocin | Eurocin | Oeemig et al. 2012 | A. montevidensis |
| Diterpene antibiotics | LL-S491β, LL-S491γ | Ellestad et al. 1972 | A. chevalieri |
| Asperglaucide | Asperglaucide | Cox et al. 1976 | A. glaucus |
| Hopane type triterpenoids | 2-Hydroxydiplopterol | Wang et al. 2009 (misidentified as A variecolor) | Aspergillus section Aspergillus species |
| Mycotoxin production by Aspergillus section Aspergillus strains checked but not confirmed in this study | |||
| Citrinin (produced by a contaminant?) | Li et al. 2006 | A. glaucus? (as Microsporum sp.) | |
| Aflatoxins (not produced) (Bachmann et al., 1979, Bachmann et al., 1982, Blaser et al., 1980, Varga et al., 2009) | Kulik and Holaday, 1966, Leitao et al., 1989, Jayraman and Kalyanasundaram, 1990, El-Kady et al., 1994, Ahmed et al., 2005, Fraga et al., 2007, Fraga et al., 2008 | A. chevalieri, A. intermedius, A. pseudoglaucus, A. ruber | |
| Sterigmatocystin (not produced) (Rank et al. 2011) | Schroeder and Kelton, 1975, Moubasher et al., 1977, Szebiotko et al., 1981, Karo and Hadlok, 1982, Soboleva and Kurmanov, 1984, El-Kady et al., 1994, Ahmed et al., 2005 | A. chevalieri, A. intermedius, A. montevidensis, A. pseudoglaucus, A. ruber | |
| Xanthocillin X (not produced) (Bachmann et al., 1979, Bachmann et al., 1982, Blaser et al., 1980) | Coveney et al. 1966 | A. glaucus | |
| Gliotoxin (not produced) (Bachmann et al., 1979, Bachmann et al., 1982, Blaser et al., 1980) | Wilkinson and Spilsbury, 1965, El-Kady et al., 1994 | A. chevalieri, A. intermedius, A. pseudoglaucus | |
| Ochratoxin A (not produced) (Bachmann et al., 1979, Bachmann et al., 1982, Blaser et al., 1980) | Chelkowski et al., 1987, El-Kady et al., 1994, Al-Julaifi, 2003 | A. glaucus, A. montevidensis, A. pseudoglaucus | |
Chevalone A-D, aszonapyrone A-B, eurochevalierine and CJ-12662 reported from Eurotium chevalieri were produced by a strain from Aspergillus section Fumigati (see Frisvad & Larsen 2016).
These compounds may have been produced by a strain of Aspergillus section Fumigati contaminating A. pseudoglaucus (Eurotium repens), as they have been found co-occurring in Aspergillus fumigatus (see Frisvad & Larsen 2016), but never in Aspergillus section Aspergillus.
Diketopiperazine dimers.
Table 8.
Extrolites found in the different species of Aspergillus section Aspergillus. Tetracyclic means compounds with a UV spectrum typical of BMS-192548 (Shu et al. 1995) or similar UV spectra.
| Species | Extrolites |
|---|---|
| Aspergillus aerius | Auroglaucin, bisanthrons, dihydroauroglaucin, echinulins, erythroglaucin, flavoglaucin, isoechinulins, neoechinulins, physcion, tetracyclic, tetrahydroauroglaucin |
| A. appendiculatus | Asperflavin, auroglaucin, bisanthrons, dihydroauroglaucin, echinulins, emodin, erythroglaucin, flavoglaucin, isoechinulins, neoechinulins, physcion, questin, questinol, tetracyclic, tetrahydroauroglaucin, “MYO” |
| A. aurantiacoflavus | Asperflavin, auroglaucin, bisanthrons, dihydroauroglaucin, echinulins, emodin, epiheveadrides, erythroglaucin, flavoglaucin, isoechinulins, neoechinulins, physcion, questin, questinol, tetracyclic, tetrahydroauroglaucin |
| A. brunneus | Asperflavin, asperentins, auroglaucin, bisanthrons, dihydroauroglaucin, echinulins, 5-farnesyl-5,7-dihydroxy-4-methylphthalide, erythroglaucin, flavoglaucin, isoechinulins, mycophenolic acid, neoechinulins, physcion, questin, tetracyclic, tetrahydroauroglaucin |
| A. caperatus | Auroglaucin, a bisanthron, dihydroauroglaucin, echinulins, epiheveadrides, flavoglaucin, isoechinulins, neoechinulins, physcion, tetrahydroauroglaucin |
| A. chevalieri | Asperflavin, auroglaucin, bisanthrons, dihydroauroglaucin, echinulins, emodin, epiheveadrides, flavoglaucin, isoechinulins, neoechinulins, physcion, questin, questinol, tetracyclic, tetrahydroauroglaucin, unique: “MYO” |
| A. cibarius | Asperflavin, auroglaucin, bisanthrons, dihydroauroglaucin, echinulins, emodin, erythroglaucin, flavoglaucin, neoechinulins physcion, tetracyclic, tetrahydroauroglaucin |
| A. costiformis | Auroglaucin, dihydroauroglaucin, echinulins, epiheveadrides, flavoglaucin, isoechinulins, neoechinulins, physcion, tetrahydroauroglaucin |
| A. cristatus | Asperflavin, auroglaucin, bisanthrons, dihydroauroglaucin, echinulins, emodin, epiheveadrides, erythroglaucin, flavoglaucin, isoechinulins, neoechinulins, physcion, tetrahydroauroglaucin, “MYO” |
| A. cumulatus | Auroglaucin, bisanthrons, dihydroauroglaucin, echinulins, emodin, erythroglaucin, flavoglaucin, isoechinulins, neoechinulins, physcion, tetracyclic, tetrahydroauroglaucin (DTO 308-D8: “canescins”; DTO 355-G9 produces another type of indolealkaloids than echinulins in addition to echinulins, similar to 12,13-dehydro-deoxybrevianamide E) |
| A. endophyticus | Auroglaucin, bisanthrones, dihydroauroglaucin, echinulins (for all species we did find preechinulin, echinulin, neoechinulin A and B), emodin, erythroglaucin, flavoglaucin, isoechinulins, neoechinulins, physcion, tetrahydroauroglaucin |
| A. glaucus | Asperflavin, auroglaucin, bisanthrons, dihydroauroglaucin, echinulins, emodin, epiheveadrides (traces), erythroglaucin, flavoglaucin, isoechinulins, neoechinulins, physcion, questin, questinol, tetracyclic, tetrahydroauroglaucin |
| A. intermedius | Asperflavin, auroglaucin, dihydroauroglaucin, echinulins, epiheveadrides, flavoglaucin, isoechinulins, LL-S491β, neoechinulins, physcion, questin, tetrahydroauroglaucin |
| A. leucocarpus | An apolar indoloterpene, echinulins, epiheveadrides, neoechinulins, “MUDI” 1-3 |
| A. levisporus | Auroglaucin, dihydroauroglaucin, echinulins, flavoglaucin, isoechinulins, neoechinulins, tetrahydroauroglaucin, unique: “WOF” 1 & 2 |
| A. mallochii | Auroglaucin, dihydroauroglaucin, echinulins, erythroglaucin, flavoglaucin, isoechinulins, neoechinulins, tetracyclic, tetrahydroauroglaucin |
| A. megasporus | Asperflavin, auroglaucin, bisanthrons, dihydroauroglaucin, echinulin, emodin, erythroglaucin, flavoglaucin, isoechinulins, neoechinulins, preechinulin, physcion, quinolactacin (A1, A2, B), tetracyclic, tetrahydroauroglaucin |
| A. montevidensis | Apolar indoloterpenes, asperflavin in few isolates, auroglaucin, dihydroauroglaucin, echinulins, epiheveadrides, flavoglaucin, isoechinulins, neoechinulins, tetrahydroauroglaucin |
| A. neocarnoyi | Asperentins, asperflavin, auroglaucin, a bisanthron, dihydroauroglaucin, echinulins, flavoglaucin, neoechinulins, questin, questinol, tetracyclic, tetrahydroauroglaucin |
| A. niveoglaucus | Asperflavin, auroglaucin, bisanthrons, dihydroauroglaucin, echinulins, emodin, erythroglaucin, flavoglaucin, mycophenolic acid (tentatively identified), neoechinulins, physcion, questin, questinol, siderin (in DTO 355-C4), tetracyclic, tetrahydroauroglaucin |
| A. osmophilus | Asperflavin, auroglaucin, flavoglaucin, dihydroauroglaucin, tetrahydroauroglaucin, echinulin and neoechinulin A. |
| A. porosus | Asperflavin, auroglaucin, bisanthrons, dihydroauroglaucin, echinulins, emodin, epiheveadrides, isoechinulins, flavoglaucin, neoechinulins, physcion, tetrahydroauroglaucin |
| A. proliferans | Asperflavin, auroglaucin, bisanthrons, dihydroauroglaucin, echinulins, emodin, epiheveadrides, erythroglaucin, flavoglaucin, isoechinulins, neoechinulins, physcion, questin, tetracyclic, tetrahydroauroglaucin |
| A. pseudoglaucus | Asperentins, asperflavin, auroglaucin, bisanthrons, dihydroauroglaucin, echinulins, erythroglaucin, 6-farnesyl-5,7-dihydroxy-4-methylphthalide, flavoglaucin, isoechinulins, mycophenolic acid, neoechinulins, physcion, questin, questinol, tetracyclic, tetrahydroauroglaucin |
| A. ruber | Auroglaucin, bisanthrons, catenarin, dihydroauroglaucin, echinulins, epiheveadrides, erythroglaucin, flavoglaucin, isoechinulins, neoechinulins, physcion, questin, questinol, tetracyclic, tetrahydroauroglaucin (Ex type of A. athecius CBS 464.65 produced breviones and no red anthraquinones) |
| A. sloanii | Auroglaucin, bisanthrons, dihydroauroglaucin, echinulins, flavoglaucin, physcion, tetracyclic, tetrahydroauroglaucin |
| A. tamarindosoli | Asperflavin, auroglaucin, bisanthrons, echinulins, emodin, dihydroauroglaucin, epiheveadrides, flavoglaucin, isoechinulins, neoechinulins, physcion, tetrahydroauroglaucin, unique: “MYO” |
| A. teporis | Echinulins, epiheveadrides, isoechinulins, neoechinulins, unique: “KYF” 1 & 2 |
| A. tonophilus | Auroglaucin, bisanthrons (few), dihydroauroglaucin, echinulins, flavoglaucin, an apolar indoloterpene, isoechinulins, neoechinulins, tetrahydroauroglaucin |
| A. xerophilus | Bisanthrons, dihydroauroglaucin, echinulins, erythroglaucin, isoechinulins, neoechinulins, physcion, sulochrin, tetracyclic, unique: “XERO” |
| A. zutongqii | Asperflavin, auroglaucin, bisanthrons, dihydroauroglaucin, echinulins, emodin, epiheveadrides, erythroglaucin, flavoglaucin, isoechinulins, neoechinulins, (tetracyclic), tetrahydroauroglaucin |
Nearly all species in sect. Aspergillus produce auroglaucins (Table 8). These heptaketides contribute to the yellow colour of the ascomata in this group. Aspergillus leucocarpus does not produce auroglaucins, partly explaining its cream to white coloured ascomata. Aspergillus teporis also does not produce auroglaucins, and this species produces less bright creamish yellow ascomata, albeit not creamish white. Aspergillus xerophilus produces a small amount of dihydroauroglaucin, but not auroglaucin, flavoglaucin and tetrahydroauroglaucin. The hexaketide siderin is recovered from one strain of A. niveoglaucus, but this kind of compound related to orlandin and kotanins is more commonly produced in sections Nigri and Clavati (Varga et al., 2007, Nielsen et al., 2009). Aspergillus pseudoglaucus is an efficient producer of the meroterpenoid mycophenolic acid and was already reported to produce the tetraketide precursor 5,7-dihydroxy-4-methylphthalide by Grove (1972a), albeit misidentified as a strain of A. flavus. Mycophenolic acid and its precursors have also been reported from sect. Aspergillus (as Eurotium spp.) by Burkin and Kononenko, 2010, Gao et al., 2011, Gao et al., 2012a, Gao et al., 2012b and Séguin et al. (2014). Epiheveadrides are detected in many species (Table 8). These nonadrides are biosynthesized from a polyketide and components from the citric acid cycle (Williams et al. 2016). They are unique to this group of Aspergilli.
Even though some extrolites from sect. Aspergillus have been claimed to be toxic (Bachmann et al., 1979, Bachmann et al., 1982, Blaser et al., 1980), these metabolites do not follow the definition of a mycotoxin. However, in higher amounts echinulins may be toxic when ingested as feed. The toxicity of echinulins and other secondary metabolites from Aspergillus sect. Aspergillus may need a re-evaluation as potential mycotoxins. The possible human toxicity of these compounds also needs to be re-evaluated. The real mycotoxins aflatoxin, sterigmatocystin, gliotoxin, citrinin, ochratoxin A could not be recovered from any of the species in sect. Aspergillus (Table 7, Table 8). In fact, the species in this group may contribute to the healthiness of fermented products such as golden tea and katsuobushi via their strong antioxidant properties of their extrolites (Ishikawa et al., 1985, Li et al., 2004a, Miyake et al., 2009, Li et al., 2009, Meng et al., 2016).
Occurrence of Aspergillus section Aspergillus species in indoor environment
Isolations from indoor environments including air, air treatment systems, dust resulted in 96 Aspergillus sect. Aspergillus strains originating from fifteen countries including Belgium, Canada, Czech Republic, France, Germany, Hungary, Mexico, Puerto Rico, South Africa, Thailand, the Netherlands, Trinidad & Tobago, Turkey, UK and USA. Strains were identified using CaM sequences, with respective GenBank numbers shown in Table 9. Fourty-three (45 %) strains were identified as A. pseudoglaucus, 20 (21 %) as A. montevidensis, and 12 (13 %) as A. chevalieri. The remaining strains were identified as A. appendiculatus, A. cibarius, A. glaucus, A. intermedius, A. leucocarpus, A. niveoglaucus, A. proliferans, A. ruber and a new species A. aerius.
Table 9.
Identification of indoor section Aspergillus species from fifteen countries.
| Species | Strain no. | Substrate | Location | CaM GenBank accession nr. |
|---|---|---|---|---|
| Aspergillus aerius | CBS 141771 = DTO 241-G7 | Air treatment system in plant production | The Netherlands | LT670991 |
| A. appendiculatus | DTO 197-F5 | Air, bakery | Tilburg, the Netherlands | LT671231 |
| A. chevalieri | DTO 080-H3 | Air, house | Stuttgart, Germany | LT671221 |
| DTO 106-E5 | Vultures enclosure (indoor) | Amsterdam, the Netherlands | LT671222 | |
| DTO 124-E8 | Air in food related factory | Ospel, the Netherlands | LT671223 | |
| DTO 130-E7 | Indoor environment | Thailand | LT671224 | |
| DTO 131-B6 | Indoor environment | Thailand | LT671225 | |
| DTO 177-B1 | Air, bakery | Heerde, the Netherlands | LT671226 | |
| DTO 177-B3 | Air, bakery | Heerde, the Netherlands | LT671227 | |
| DTO 268-B7 | Houst dust | Mexico | LT671229 | |
| DTO 266-F8 | Houst dust | Thailand | LT671230 | |
| EMSL No. 2223 | Air, hospital | Fairfax, VA, USA | LT671218 | |
| EMSL No. 56 | Indoor air | California, USA | LT671219 | |
| EMSL No. 2871 | Indoor air, basement | Denver, CO, USA | LT671220 | |
| A. cibarius | DTO 123-E7 | Air, office | Zutphen, the Netherlands | LT671232 |
| DTO 124-B9 | Air in food related factory | Ospel, the Netherlands | LT671233 | |
| DTO 197-F6 | Air, bakery | Tilburg, the Netherlands | LT671234 | |
| A. glaucus | EMSL No. 2529 | Air, office | Puerto Rico | LT671071 |
| DTO 155-G4 | Indoor, paper | The Netherlands | LT671257 | |
| EMSL No. 3317 = CCF 5382 = DTO 355-H2 | Indoor air, bedroom | NY, USA | LT671074 | |
| A. intermedius | CCF 5377 = DTO 355-G5 | Air, surgical operating room | Prague, Czech Republic | LT671080 |
| A. leucocarpus | DTO 357-A2 = KAS 7576 | Houst dust | Canada | LT671089 |
| A. montevidensis | DTO 008-H7 = CBS 119376 | Indoor environment | Germany | LT671235 |
| DTO 072-E7 | Indoor, archive | Amsterdam, the Netherlands | LT671236 | |
| DTO 108-F4 | Indoor environment | France | LT671237 | |
| DTO 123-D7 | Air, office | Zutphen, the Netherlands | LT671238 | |
| DTO 126-A3 | Swab sample, kitchen cabinet drawer next to sink | The Netherlands | LT671239 | |
| DTO 146-E3 | Indoor environment | Hungary | LT671240 | |
| DTO 146-E4 | Indoor environment | Hungary | LT671241 | |
| DTO 146-E6 | Indoor environment | Hungary | LT671242 | |
| DTO 147-E4 | Indoor environment | Hungary | LT671243 | |
| DTO 177-A8 | Air, bakery | Heerde, the Netherlands | LT671244 | |
| DTO 177-A9 | Air, bakery | Heerde, the Netherlands | LT671245 | |
| DTO 177-B2 | Air, bakery | Heerde, the Netherlands | LT671246 | |
| DTO 177-B6 | Air, bakery | Heerde, the Netherlands | LT671247 | |
| DTO 177-B7 | Air, bakery | Heerde, the Netherlands | LT671248 | |
| DTO 299-A2 | Indoor hospital air | Turkey | LT671249 | |
| DTO 180-B6 | House dust | South Africa | LT671250 | |
| DTO 267-H2 | House dust | Thailand | LT671251 | |
| EMSL No. 1589 | Air, green house | Delaware, USA | LT671252 | |
| EMSL No. 2730 | Black HEPA filter | Edwardsville, IL, USA | LT671254 | |
| EMSL No. 2934 = CCF 5379 = DTO 355-H3 | Indoor air, bedroom | Mahanoy City, PA, USA | LT671098 | |
| A. niveoglaucus | DTO 177-B4 | Air, bakery | Heerde, the Netherlands | LT671255 |
| IHEM 1811 = DTO 355-C3 | Indoor air | Namur, Belgium | LT671116 | |
| EMSL No. 2211 | Air, bathroom | Great Falls, MT, USA | LT671113 | |
| A. proliferans | DTO 124-C8 | Air in food related factory | Ospel, the Netherlands | LT671256 |
| DTO 197-F7 | Air, bakery | Tilburg, the Netherlands | LT671258 | |
| DTO 197-F8 | Air, bakery | Tilburg, the Netherlands | LT671259 | |
| DTO 331-D1 | Air, house | Noordwijk, the Netherlands | LT671260 | |
| EMSL No. 2207 = CCF 5395 = DTO 355-H5 | Air of living room | Yardley, PA, USA | LT671149 | |
| EMSL No. 2791 = CCF 5392 = DTO 355-H6 | Indoor air, basement | Troy, NY, USA | LT671152 | |
| A. pseudoglaucus | DTO 011-E9 | Indoor air | Loosdrecht, the Netherlands | LT671264 |
| DTO 244-I1 | Houst dust | UK | LT671265 | |
| DTO 244-I7 | Houst dust | UK | LT671266 | |
| DTO 039-F5 | Indoor environment from mortel (cement) | Düsseldorf, Germany | LT671267 | |
| DTO 072-E6 | Indoor, archive | Amsterdam, the Netherlands | LT671268 | |
| DTO 087-G6 | Air in warehouse, Citronas | The Netherlands | LT671269 | |
| DTO 106-D1 | Elephants enclosure (indoor) | Amsterdam, the Netherlands | LT671270 | |
| DTO 106-E2 | Zebra enclosure (indoor) | Amsterdam, the Netherlands | LT671271 | |
| DTO 115-F5 | Indoor | Hungary | LT671272 | |
| DTO 117-F9 | Indoor, archive | Giessenlanden, the Netherlands | LT671273 | |
| DTO 123-D8 | Air, office | Zutphen, the Netherlands | LT671274 | |
| DTO 123-I2 | Air, factory | Kerkrade, the Netherlands | LT671275 | |
| DTO 124-D3 | Air in food related factory | Ospel, the Netherlands | LT671276 | |
| DTO 126-A2 | Swab sample, kitchen cabinet drawer next to sink | The Netherlands | LT671277 | |
| DTO 147-B6 | Indoor environment | Hungary | LT671278 | |
| DTO 147-D1 | Indoor environment | Hungary | LT671279 | |
| DTO 147-H9 | Indoor environment | Hungary | LT671280 | |
| DTO 177-B5 | Air, bakery | Heerde, the Netherlands | LT671281 | |
| DTO 241-H5 | Air treatment system in production plant | Goes, the Netherlands | LT671282 | |
| EMSL No. 2474 = CCF 5387 = DTO 355-I4 | Indoor air, basement | Piscataway, NJ, USA | LT671167 | |
| EMSL No. 2779 = CCF 5389 = DTO 355-I3 | Air in front of air conditioning vent | Melbourne, FL, USA | LT671161 | |
| EMSL No. 1022 | Indoor air of home | New Jersey, USA | LT671283 | |
| EMSL No. 1415 | Indoor air of home | Massachusetts, USA | LT671284 | |
| EMSL No. 1643 | Indoor air of hospital | Alabama, USA | LT671285 | |
| EMSL No. 1918 | Air, living room | New York, NY, USA | LT671286 | |
| EMSL No. 1919 | Air, bedroom | Fort Salonga, NY, USA | LT671287 | |
| EMSL No. 1966 | Air, hospital | New York, NY, USA | LT671288 | |
| EMSL No. 2222 | Air, bedroom | Cinnaminson, NJ, USA | LT671289 | |
| EMSL No. 1245 | Air, home | New Jersey, USA | LT671290 | |
| EMSL No. 1246 | Air, home | New Jersey, USA | LT671291 | |
| EMSL No. 2130 | Air, hospital | Trinidad & Tobago | LT671292 | |
| EMSL No. 2472 | Air, basement | Piscataway, NJ, USA | LT671293 | |
| EMSL No. 2473 | Air, basement | Piscataway, NJ, USA | LT671294 | |
| EMSL No. 2475 | Air, basement | Piscataway, NJ, USA | LT671295 | |
| EMSL No. 2832 | Carpet dust, Harker Heights Trails | TX, USA | LT671296 | |
| EMSL No. 2834 | Air, hospital | Chicago, IL, USA | LT671297 | |
| EMSL No. 2844 | Swab, bedroom | Norman, OK, USA | LT671298 | |
| EMSL No. 2845 | Swab, bedroom | Norman, OK, USA | LT671299 | |
| EMSL No. 2860 | Air, living room | Bowling Green, KY, USA | LT671300 | |
| EMSL No. 2861 | Air, living room | Bowling Green, KY, USA | LT671301 | |
| EMSL No. 2930 | Air, living room | Big Rapids, RI, USA | LT671302 | |
| EMSL No. 1780 = CCF 5388 = DTO 355-I2 | House dust | Pennsylvania, USA | LT671158 | |
| EMSL No. 2809 = CCF 5386 | Indoor air, office | Endicott, NY, USA | LT671164 | |
| A. ruber | DTO 146-E2 | Indoor environment | Hungary | LT671261 |
| DTO 267-H3 | House dust | Thailand | LT671262 |
Members of sect. Aspergillus are able to grow on all types of organic materials at low moisture levels, therefore this group of fungi is frequently reported from the indoor environment (Samson et al., 2010, Šimonovičová et al., 2015, Thrasher, 2016, Visagie et al., 2014a, Visagie et al., 2017). Since their wide distribution and environmental adaptation, sect. Aspergillus species were used as biosensor fungi to assess indoor climate and predict hidden moisture damage in homes (Abe et al., 1996, Baudisch et al., 2009). Samson et al. (2010) listed four common sect. Aspergillus species in indoor environment including A. montevidensis (= E. amstelodami), A. chevalieri (= E. chevalieri), A. ruber (= E. rubrum) and A. glaucus (= E. herbariorum). Visagie et al. (2014a) reported six species including A. ruber, A. proliferans, A. montevidensis, A. pseudoglaucus, A. sloanii and A. chevalieri from house dust samples, and more recently, Visagie et al. (2017) reported another nine species from Canadian and Hawaiian dust. In the current study A. pseudoglaucus, A. montevidensis and A. chevalieri represented 78 % of all isolates. Phenotypically, these indoor species are very similar. Aspergillus pseudoglaucus is similar to A. proliferans and A. ruber, while A. montevidensis and A. chevalieri only bear small differences in ascospore ornamentation, and can be confused with others in the A. chevalieri clade (such as A. intermedius and A. caperatus). Thus molecular identification especially CaM instead of ITS is recommended for accurate identification. Eurotium amstelodami and E. repens are two of the most encountered names in indoor sect. Aspergillus species (Samson et al., 2010, Šimonovičová et al., 2015, Thrasher, 2016). The names A. montevidensis (= E. amstelodami) and A. pseudoglaucus (= E. repens), respectively, were chosen based on priority and new nomenclature rules (McNeill et al., 2012, Hubka et al., 2013a). To keep the consistent species concepts which facilitate comparable research, this treatment is followed in this study.
Key to the most common section Aspergillus species from the indoor environments
| 1a) Ascospores with high crests (0.5–1 μm) | 2 |
| 1b) Ascospores with low crests (< 0.5 μm) or crests lacking | 4 |
| 2a) Ascospores with smooth or faintly roughened convex surface | 3 |
| 2b) Ascospores with rugulose convex surface | A. montevidensis |
| 3a) Ascospores small, with spore bodies measuring 3.5–5.5 × 3–4 μm | A. chevalieri |
| 3b) Ascospores large, with spore bodies measuring 5.5–7.5 × 3.5–6 μm | A. glaucus |
| 4a) Ascospores with low crests (< 0.5 μm) | 5 |
| 4b) Ascospores lack crests | 6 |
| 5a) Conidia small, 4–7 × 3.5–5.5 μm | A. cibarius |
| 5b) Conidia large, (6–)8–13.5 × 4–9 μm | A. niveoglaucus |
| 6a) Ascospores with furrow present or pronounced | 7 |
| 6b) Ascospores with furrow absent or showing as a trace | A. pseudoglaucus |
| 7a) Grows well on M60Y at 37 °C | A. ruber |
| 7b) Does not grow on M60Y at 37 °C | A. proliferans |
Taxonomy
Aspergillus section Aspergillus
Synonyms: Eurotium Link, Mag. Ges. Naturf. Freunde Berlin 3: 31, t. 2:44. 1809.
Pyrobolus Kuntze, Revis. Gen. Pl. 2: 868. 1891. fide Kuntze 1891, Dict. Fungi 10th ed.
Edyuillia Subram., Curr. Sci. 41: 756. 1972, fide Samson 1979.
Gymnoeurotium Malloch & Cain, Canad. J. Bot. 50: 2619. 1972, fide Samson 1979, Benny & Kimbrough 1980.
Conidiophores with smooth stipes, hyaline or light brown. Vesicles globose to subglobose, uniseriate, fertile over two thirds to entire surface. Phialides flask-shaped. Conidia globose, subglobose to ellipsoidal, microtuberculate, tuberculate to lobate-reticulate. Ascomata eurotium-like, cleistothecial, superficial, yellow or rarely white to cream yellow, globose to subglobose. Asci 8-spored, globose to subglobose. Ascospores one-celled, hyaline, lenticular, in surface view globose to subglobose, generally showing an equatorial furrow with or without crests, spore bodies smooth or with different degree of rough ornamentation. Xerophilic and osmophilic, growing optimally on substrates containing high concentrations of sugar or salt.
Typus: Aspergillus glaucus (L.) Link, Mag. Ges. Naturf. Freunde Berlin 3: 16. 1809.
Notes: The genus Pyrobolus was considered as synonym of Eurotium (Kuntze 1891). The genera Edyuillia and Gymnoeurotium were both based on Aspergillus athecius Raper & Fennell, Samson (1979) suspected the type culture (CBS 464.65) of A. athecius had lost its ability to produce ascomata and represented an atypical form of Eurotium, thus regarded these two genera as synonymous with Eurotium. This was further proved by phylogenetic analyses of Hubka et al. (2013a), where A. athecius was synonymized with A. ruber.
Clade classification in section Aspergillus
Aspergillus ruber clade
Most members of this clade produce non-crested ascospores or ascospores with reduced crests, the only exception is A. cumulatus, which produces irregular, low (<0.5 μm) crests. All species in this clade cannot grow on CY20S at 37 °C, four species (A. appendiculatus, A. cumulates, A. mallochii, A. sloanii) cannot grow on M60Y at 37 °C. Most species except A. appendiculatus can grow on CY20S at 25 °C, all species grow rapidly on M60Y at 25 °C.
Accepted species:
Aspergillus appendiculatus Blaser 1975, Sydowia 28: 38. [MB309209].
Aspergillus cumulatus D.H. Kim & S.B. Hong, J. Microbiol. Biotechnol. 24: 335. 2014. [MB807118].
Aspergillus mallochii Visagie, Yilmaz & Seifert, MycoKeys 19: 16. 2017. [MB819025].
Aspergillus pseudoglaucus Blochwitz, Ann. Mycol. 27: 207. 1929. [MB275429].
Aspergillus ruber (Jos. König et al.) Thom & Church, Aspergillus: 112. 1926. [MB490579].
Aspergillus sloanii Visagie, Hirooka & Samson, Stud. Mycol. 78: 108. 2014. [MB809194].
Aspergillus tonophilus Ohtsuki, Bot. Mag. (Tokyo) 75: 438. 1962. [MB326663].
Aspergillus zutongqii A.J. Chen, Frisvad & Samson, this study [MB818739].
Aspergillus glaucus clade
Members of this clade produce non-crested ascospores or ascospores with low crests (<0.5 μm) or irregular crests measuring 0.5–1 μm. All species in this clade cannot grow on CY20S and M60Y at 37 °C. Most species grow moderately on CY20S and grow rapidly on M60Y at 25 °C, except A. neocarnoyi grows restrictedly on CY20S at 25 °C (3–5 mm after 7 d).
Accepted species:
Aspergillus aerius A.J. Chen, Frisvad & Samson, this study [MB818731].
Aspergillus aurantiacoflavus Hubka, A.J. Chen, Jurjević & Samson, this study [MB818732].
Aspergillus brunneus Delacr., Bull. Soc. Mycol. France 9: 185. 1893. [MB204832].
Aspergillus glaucus (L.) Link, Mag. Ges. Naturf. Freunde Berlin 3: 16. 1809. [MB161735].
Aspergillus levisporus Hubka, A.J. Chen, Jurjević & Samson, this study [MB818735].
Aspergillus megasporus, Visagie, Yilmaz & Seifert, MycoKeys 19: 17. 2017. [MB819028].
Aspergillus niveoglaucus Thom & Raper, U.S.D.A. Misc. Pub. 426: 35. 1941. [MB120985].
Aspergillus neocarnoyi Kozak., Mycol. Pap. 161: 63. 1989. [MB127756].
Aspergillus proliferans G. Sm., Trans. Brit. Mycol. Soc. 26: 26. 1943. [MB284312].
Aspergillus chevalieri clade
Members of this clade produce ascospores with high crests (≥ 0.5 μm). All species in this clade can grow on M60Y at 37 °C. Most species except A. caperatus and A. costiformis can grow on CY20S at 37 °C. All species grow rapidly on CY20S and M60Y at 25 °C.
Accepted species:
Aspergillus caperatus A.J. Chen, Frisvad & Samson, this study [MB818733].
Aspergillus chevalieri (L. Mangin) Thom & Church, The Aspergilli: 111. 1926. [MB292839].
Aspergillus costiformis H.Z. Kong & Z.T. Qi, Acta Mycol. Sin. 14: 10. 1995. [MB363444].
Aspergillus cristatus Raper & Fennell, Gen. Aspergillus: 169. 1965. [MB326622].
Aspergillus intermedius Blaser, Sydowia 28: 41. 1975. [MB309226].
Aspergillus montevidensis Talice & Mackinnon, Compt. Rend. Soc. Biol. Fr. 108: 1007. 1931. [MB309231].
Aspergillus porosus A.J. Chen, Frisvad & Samson, this study [MB818736].
Aspergillus xerophilus clade
Members of this clade produce ascospores with low crests (≤ 0.5 μm), cannot grow on CYA, MEA, CY20S, while grow rapidly on M60Y. Aspergillus osmophilus grows rapidly on M60Y at 37 °C, while A. xerophilus does not grow under this condition.
Accepted species:
Aspergillus osmophilus Asgari & Zare, Mycoscience 55: 58. 2013. [MB803278].
Aspergillus xerophilus Samson & Mouch., Antonie van Leeuwenhoek 41: 348. 1975. [MB309251].
Other species:
Aspergillus cibarius S.B. Hong & Samson, J. Microbiol. 50: 713. 2012. [MB800861].
Aspergillus endophyticus Hubka, A.J. Chen, & Samson, this study [MB818734].
Aspergillus leucocarpus Hadlok & Stolk, Antonie van Leeuwenhoek 35: 9. 1969. [MB326642].
Aspergillus tamarindosoli A.J. Chen, Frisvad & Samson, this study [MB818737].
Aspergillus teporis A.J. Chen, Frisvad & Samson, this study [MB818738].
Species Descriptions
Aspergillus aerius A.J. Chen, Frisvad & Samson, sp. nov. MycoBank MB818731. Fig. 18.
Fig. 18.
Aspergillus aerius CBS 141771T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Etymology: Name refers to its origin, isolated from air treatment system.
Diagnosis: Large (6.5–8 × 4.5–6 μm), smooth ascospores with roughness along equatorial ridges, tuberculate conidia measuring (5–)10–13 × 6–10 μm.
Typus: The Netherlands, air treatment system in production plant, 2013, isolated by J. Houbraken (holotype CBS H-22823, culture ex-type: CBS 141771 = DTO 241-G7 = IBT 34446).
ITS barcode: LT670916. (Alternative markers: BenA = LT670990; CaM = LT670991; RPB2 = LT670992).
Colony diam, 7 d (mm): CYA 10–12; MEA 7–10; CY20S 17–20; CY20S 30 °C 14–15; CY20S 37 °C No growth; M40Y 65–66; M60Y >75; M60Y 30 °C >75; M60Y 37 °C No growth; CYAS 35–36; DG18 40–44; MEA10S 63–65.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium pale luteous (11) to sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse pale luteous (11). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) to orange (7); texture floccose; sporulation moderately dense; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse ochreous (44). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) to orange (7); texture floccose; sporulation moderately dense; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse ochreous (44). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation dense; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse buff (45). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse amber (47). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium sulphur yellow (15); margins entire; texture floccose; sporulation moderately dense; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse ochreous (44).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 190–275 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies smooth, 6.5–8 × 4.5–6 μm, rough along equatorial ridges, in side view lenticular, furrow present, crests absent. Conidiophores with smooth stipes, hyaline or light brown, 500–1 000 × 7–15.5 μm. Vesicles globose to subglobose, 26–41 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 7.5–12.5 × 5–8 μm. Conidia globose, subglobose to ellipsoidal, tuberculate, (5–)10–13 × 6–10 μm.
Distinguishing characters: The large ascospores of A. aerius resemble those of A. brunneus, but A. brunneus produces larger conidia, that are infrequently ellipsoidal (8–15 × 8–13 μm).
Aspergillus appendiculatus Blaser 1975, Sydowia 28: 38. MycoBank MB309209. Fig. 19.
Fig. 19.
Aspergillus appendiculatus CBS 374.75T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Synonyms: Eurotium appendiculatum Blaser, Sydowia 28: 38. 1975.
Aspergillus aridicola H.Z. Kong & Z.T. Qi, Acta Mycol. Sin. 14: 88. 1995.
Eurotium aridicola H.Z. Kong & Z.T. Qi, Acta Mycol. Sin. 14: 88. 1995.
Typus: ZT 8286, holotype. Culture ex-type: CBS 374.75 = IMI 278374 = FRR 2793 = JCM 1566 = PIL 588 = IBT 34507.
ITS barcode: HE615132. (Alternative markers: BenA = HE801333; CaM = HE801318; RPB2 = HE801307).
Colony diam, 7 d (mm): CYA No growth; MEA No growth; CY20S No growth; CY20S 30 °C No growth; CY20S 37 °C No growth; M40Y 50–52; M60Y 50–59; M60Y 30 °C 44–49; M60Y 37 °C No growth; CYAS 19–20; DG18 35–38; MEA10S 26–28.
Colony characters: M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse light citrine green (67). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse sulphur yellow (15) in the centre, citrine green (67) in the edge. MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium white; margins entire; texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse sulphur yellow (15).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 100–225 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies slightly rough, 5–7.5 × 4–5.5 μm, in side view lenticular, furrow absent or showing as a trace, crests with filiform appendages or petaliform, petals 1–1.5 μm wide at highest parts. Conidiophores with smooth stipes, hyaline or light brown, 800–2 000 × 7–12(–14.5) μm. Vesicles globose to subglobose, 30–64 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 8–16 × 4.5–7.5 μm. Conidia globose, subglobose to ellipsoidal, tuberculate, 5–10(–12) × 5–7(–8.5) μm.
Distinguishing characters: Aspergillus appendiculatus is typically characterized by petaliform crests on ascospores. Similar ascospores are also produced by A. mallochii, which are smaller in size (4–6 × 3–5 μm).
Additional materials examined: Canada, House dust, 2015, isolated by C.M. Visagie, DTO 357-A3 = KAS 7579. China, Tibet, sheep dung, isolated by H.Z. Kong & Z.T. Qi, CBS 101746 = CGMCC 3.04673 (AS 3.4673).
Aspergillus aurantiacoflavus Hubka, A.J. Chen, Jurjević & Samson, sp. nov. MycoBank MB818732. Fig. 20.
Fig. 20.
Aspergillus aurantiacoflavus CBS 141930T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Etymology: Name refers to its orange and yellow colony, aurantiacus = orange, flavus = yellow.
Diagnosis: Orange and yellow colony, verruculose ascospores measuring 4–5.5 × 3–5 μm.
Typus: USA, CA, San Diego, baby carrier backpack, 2015, isolated by Ž. Jurjević (holotype CBS H-22827, culture ex-type: CBS 141930 = EMSL No. 2903 = CCF 5393 = DTO 355-I1 = IBT 34485).
ITS barcode: LT670917. (Alternative markers: BenA = LT670993; CaM = LT670994; RPB2 = LT670995).
Colony diam, 7 d (mm): CYA 2–3; MEA 2–3; CY20S 23–25; CY20S 30 °C No growth; CY20S 37 °C No growth; M40Y 65–70; M60Y 70–>75; M60Y 30 °C >75; M60Y 37 °C No growth; CYAS 38–40; DG18 44–45; MEA10S 44–45.
Colony characters: CY20S 25 °C, 7 d: Colonies low to moderately deep, plane; margins entire; mycelium sulphur yellow (15) and orange (7); texture floccose; sporulation absent to sparse; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse pale luteous (11) or buff (45). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) and orange (7); texture floccose; sporulation sparse; conidia en masse pale green (19) to greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44) or orange (7) at centre, ochreous (44) at edge. M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) and orange (7); texture floccose; sporulation sparse to moderately dense; conidia en masse pale green (19) to greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) and orange (7); texture floccose; sporulation absent to moderately dense; conidia en masse pale green (19) to greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44) or orange (7) at centre, ochreous (44) at edge. DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) and orange (7); texture floccose; sporulation sparse to moderately dense; conidia en masse pale green (19) to greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44) or orange (7) at centre, ochreous (44) at edge. MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium sulphur yellow (15) and orange (7); margins entire; texture floccose; sporulation absent to moderately dense; conidia en masse pale green (19) to greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44) or orange (7) at centre, ochreous (44) at edge.
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 110–250 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies verruculose, 4–5.5 × 3–5 μm, in side view lenticular, furrow present, crests irregular, <0.5 μm. Conidiophores with smooth stipes, hyaline or light brown, 250–800 × 7.5–12 μm. Vesicles globose to subglobose, 30–45 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 6–11 × 3.5–6.5 μm. Conidia globose to subglobose, tuberculate, 5–9 × 4–7 μm.
Distinguishing characters: Phylogenetically A. aurantiacoflavus is closely related to A. proliferans and A. glaucus, but A. proliferans produces non-crested ascospores; A. glaucus produces larger ascospores (5.5–7.5 × 3.5–6 μm).
Additional materials examined: USA, IL, Chicago, rubber toy import from China, 2015, isolated by Ž. Jurjević, CCF 5562 = EMSL No. 2690, CCF 5563 = EMSL No. 2691, CCF 5564 = EMSL No. 2692, EMSL No. 2693 = CCF 5391 = DTO 355-H7, CCF 5565 = EMSL No. 2694. USA, New Jersey, Cherry Hill, cake spread, 2015, isolated by Ž. Jurjević, EMSL No. 3024 = CCF 5394 = DTO 355-H9.
Aspergillus brunneus Delacr., Bull. Soc. Mycol. France 9: 185. 1893. MycoBank MB204832. Fig. 21.
Fig. 21.
Aspergillus brunneus CBS 112.26T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Synonyms: Eurotium echinulatum Delacr., Bull. Soc. Mycol. France 9: 266. 1893.
Aspergillus echinulatus (Delacr.) Thom & Church, The Aspergilli: 107. 1926.
Aspergillus medius R. Meissn., Bot. Z.: 356. 1897.
Eurotium medium R. Meissn., Bot. Z.: 356. 1897.
Eurotium verruculosum Vuill. Bull. Soc. Mycol. France 34: 83. 1918.
Typus: IMI 211378, epitype (Hubka et al. 2013a). Culture ex-type: CBS 112.26 = CBS 524.65 = IBT 5341 = NRRL 131 = NRRL 134 = ATCC 1021 = IFO 5862 = IMI 211378 = QM 7406 = Thom 4481 = Thom 5633.4 = WB 131.
ITS barcode: EF652060. (Alternative markers: BenA = EF651907; CaM = EF651998; RPB2 = EF651939).
Colony diam, 7 d (mm): CYA 11–12; MEA 3–5; CY20S 30–34; CY20S 30 °C No growth; CY20S 37 °C No growth; M40Y 65–72; M60Y >75; M60Y 30 °C 60–65; M60Y 37 °C No growth; CYAS 26–27; DG18 60–61; MEA10S 48–52.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse fulvous (43). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium orange (7), later turn into bay (6); texture floccose; sporulation dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse ochreous (44). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium orange (7); texture floccose; sporulation dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse ochreous (44). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium orange (7); texture floccose; sporulation dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse ochreous (44). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium orange (7); texture floccose; sporulation dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse fulvous (43). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium orange (7); margins entire; texture floccose; sporulation dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse fulvous (43).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 110–240 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies rough along equatorial ridges, 7–10 × 6–8 μm, in side view lenticular, furrow present, crests irregular, <0.5 μm. Conidiophores with smooth stipes, hyaline or light brown, 700–1 200 × 7–18 μm. Vesicles globose to subglobose, 32–58 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 10–18.5 × 7–12.5 μm. Conidia globose to subglobose, tuberculate, 8–15 × 8–13 μm.
Distinguishing characters: Aspergillus brunneus is close to A. neocarnoyi and A. osmophilus in ascospore size and ornamentation, but the latter two are more xerophilic, and show no growth on MEA and CYA. In addition, A. brunneus grows faster on CY20S.
Additional materials examined: Canada, house dust, 2015, isolated by C.M. Visagie, DTO 357-A1 = KAS7575. Canada, Manitoba, unknown source, isolated by M. Desjardins, DTO 197-B3 = CBS 117328. Unknown source, isolated by G. Smith, NRRL 133 = CCF 5586. Unknown source, isolated by W. McRae, NRRL 124 = CBS 113.27 = CCF 5585.
Aspergillus caperatus A.J. Chen, Frisvad & Samson, sp. nov. MycoBank MB818733. Fig. 22.
Fig. 22.
Aspergillus caperatus CBS 141774T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Etymology: Name refers to the wrinkled ornamentation on conidia.
Diagnosis: Verruculose to rugulose ascospores with crests measuring 0.5–1 μm, lobate-reticulate conidia, no growth on CY20S at 37 °C.
Typus: South Africa, Robben Island, soil, 2015, collected by P. Crous, isolated by M. Meijer (holotype CBS H-22825, culture ex-type: CBS 141774 = DTO 337-E6 = IBT 34451).
ITS barcode: LT670922. (Alternative markers: BenA = LT671008; CaM = LT671009; RPB2 = LT671010).
Colony diam, 7 d (mm): CYA 19–20; MEA 14–15; CY20S 55–56; CY20S 30 °C 52–53; CY20S 37 °C No growth; M40Y >75; M60Y >75; M60Y 30 °C >75; M60Y 37 °C >75; CYAS 41–44; DG18 49–50; MEA10S 58–63.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12) to sulphur yellow (15). M40Y 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12). M60Y 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse sulphur yellow (15). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse sulphur yellow (15). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium sulphur yellow (15); margins entire; texture floccose; sporulation sparse; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 130–220 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies verruculose to rugulose, 3.5–4.5 × 2.5–4 μm, in side view lenticular, furrow pronounced, crests 0.5–1 μm. Conidiophores with smooth stipes, hyaline or light brown, 250–500 × 6.5–9(–12) μm. Vesicles globose to subglobose, 26–45 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 7.5–12 × 4–7.5 μm. Conidia globose to subglobose, lobate-reticulate, 3.5–5.5 × 3.5–4.5 μm.
Distinguishing characters: Phylogenetically A. caperatus is closely related to A. montevidensis, A. intermedius and A. porosus, but A. montevidensis produces larger ascospores (4–6 × 3–4.5 μm), A. intermedius produces microtuberculate conidia and A. porosus is characterized by pitted ascospores. In addition, A. caperatus does not grow on CY20S at 37 °C compared to other three species.
Aspergillus chevalieri (L. Mangin) Thom & Church, The Aspergilli: 111. 1926. MycoBank MB292839. Fig. 23.
Fig. 23.
Aspergillus chevalieri CBS 522.65T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Synonyms: Eurotium chevalieri L. Mangin, Ann. Sci. Nat., Bot.: 361. 1909.
Aspergillus chevalieri var. multiascosporus Nakaz. et al., J. Agr. Chem. Soc. Japan 10: 135–192. 1934.
Aspergillus allocotus Bat. & H. Maia, Anais Soc. Biol. Pernambuco 15: 181. 1957.
Aspergillus equitis Samson & W. Gams, Advances in Penicillium and Aspergillus Systematics: 36. 1985.
Typus: IMI 211382, neotype (Samson & Gams 1985). Culture ex-type: CBS 522.65 = NRRL 78 = ATCC 16443 = IMI 211382 = NRRL A-7803 = Thom 4125.3 = WB 78 = IBT 5680.
ITS barcode: EF652068. (Alternative markers: BenA = EF651911; CaM = EF652002; RPB2 = EF651954).
Colony diam, 7 d (mm): CYA 17–25; MEA 17–27; CY20S 23–67; CY20S 30 °C 23–60; CY20S 37 °C 3–33; M40Y 50–>75; M60Y 60–>75; M60Y 30 °C 55–>75; M60Y 37 °C >75; CYAS 23–55; DG18 27–45; MEA10S 40–52.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium straw (46) to sulphur yellow (15) to orange (7); texture velvety; sporulation sparse to moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44) or fulvous (43) fading into sulphur yellow (15). M40Y 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium sulphur yellow (15) and white or orange (7); texture floccose; sporulation sparse to moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse fulvous (43) or luteous (12) or ochreous (44). M60Y 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium sulphur yellow (15) and white or orange (7); texture floccose; sporulation sparse to moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12) to ochreous (44). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) or white or grey olivaceous (107) or orange (7); texture floccose; sporulation sparse to moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12) to ochreous (44) fading into sulphur yellow (15) or luteous (12). DG18 25 °C, 7 d: Colonies moderately deep, plane or slightly sulcate; margins entire; mycelium sulphur yellow (15) and white or orange (7); texture floccose; sporulation sparse to moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12) or ochreous (44) fading into sulphur yellow (15) or rust (39). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium sulphur yellow (15) and white or orange (7); margins entire; texture floccose; sporulation sparse to moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12) or ochreous (44) or fulvous (43).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 100–250 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies smooth to slightly verruculose, 3.5–5.5 × 3–4 μm, in side view lenticular, furrow present, crests 0.5–1 μm. Conidiophores with smooth stipes, hyaline or light brown, 200–1 000 × 6–12 μm. Vesicles globose to subglobose, 23–47 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 5.5–7.5(–10) × 3–5 μm. Conidia globose, subglobose to ellipsoidal, tuberculate to lobate-reticulate, 3–4(–6) × 2.5–3.5(–5) μm.
Distinguishing characters: Phylogenetically A. chevalieri is closely related to A. cristatus and A. costiformis, but A. cristatus produces verruculose to rugulose ascospores, while A. costiformis produces large rugulose ascospores (5.5–7 × 5–6.5 μm). Morphologically, A. chevalieri is close to A. intermedius and A. caperatus in ascospore size and ornamentation, but A. intermedius produces microtuberculate conidia, A. caperatus produces mainly globose and averagely larger conidia (3.5–5.5 × 3.5–4.5 μm).
Additional materials examined: Brazil, corn kernels, 2008, isolated by J. Houbraken, DTO 061-A2. China, Guizhou, liquor starter, CGMCC 3.06736, CGMCC 3.06722. China, unknown source, CGMCC 3.01302, CGMCC 3.01303, CGMCC 3.12591, CGMCC 3.01299, CGMCC 3.01301. China, Beijing, unknown source, CGMCC 3.06135, CGMCC 3.06136. China, Yunnan, moldy weeds, CGMCC 3.06491. China, Yunnan, moldy bamboo, CGMCC 3.06490. China, Ningxia, soil, CGMCC 3.06133. China, Yunnan, soil under corn, CGMCC 3.06487. China, Yunnan, soil, CGMCC 3.06489. China, Guizhou, liquor starter, CGMCC 3.06753. China, Beijing, soil, CGMCC 3.06134. China, Beijing, feed, CGMCC 3.07889. China, Tibet, soil, 2001, CGMCC 3.06132 = DTO 348-G5. China, Yunnan, moldy peel, 2001, CGMCC 3.06492 = DTO 348-H3. Czech Republic, Brno, rice, 1999, isolated by V. Ostrý, CCF 3291 = DTO 355-B6. Czech Republic, Prague, semolina, 1979, isolated by V. Muzikář, CCF 1676 = DTO 355-B7. Czech Republic, semolina, 1979, isolated by V. Muzikář, CCF 1663. Czech Republic, Brno, seeds of Carum carvi, 2000, isolated by V. Ostrý, CCF 3211. India, keratitis, CBS 123900. Japan, unknown source, isolated by R. Nakazawa, CBS 113.34 = NRRL 88 = WB 88 = DTO 196-H7 (Isotype of Aspergillus chevalieri var. multiascosporus). Madagascar, soil, 2008, isolated by J. Houbraken, CBS 141769 = DTO 088-D7. Madagascar, soil, 2008, isolated by J. Houbraken, DTO 092-D3. Portugal, unknown source, CBS 126335. South Korea, soybeans, 2012, isolated by D.H. Kim, CCF 4788 = KACC 47145 = DTO 355-B8. Thailand, Hua Hin, soil under tamarind, 2007, isolated by R.A. Samson & J. Houbraken, DTO 054-A9. The Netherlands, Quail bedding, 2014, isolated by M. Meijer, DTO 316-G5. The Netherlands, milk powder, 2016, isolated by J. Houbraken, DTO 346-C5. The Netherlands, animal feed kernels, 2016, isolated by J. Houbraken, DTO 346-B8. The Netherlands, garlic butter, isolated by J. Houbraken, DTO 239-H5. The Netherlands, animal feed kernels, 2016, isolated by J. Houbraken, DTO 346-B7. USA, Indiana, Indianapolis, unknown source, isolated by Dr. Adams, NRRL 79. USA, culture contamination, isolated by D.I. Fennell, NRRL 4755. USA, CA, child carrier, 2015, isolated by Ž. Jurjević, EMSL No. 2739, EMSL No. 2768. USA, AZ, Tempe, office chair, 2015, isolated by Ž. Jurjević, EMSL No. 2931. Unknown source, isolated by S. Suhendriani, DTO 238-E3.
Notes: Raper & Fennell (1965) indicated that Aspergillus chevalieri var. multiascosporus showed definitely identical colony character of A. chevalieri, and included it with A. chevalieri. Hubka et al. (2013a) synonymized A. chevalieri var. multiascosporus with A. chevalieri and our morphological observation and molecular data (CaM) supported this treatment. Aspergillus allocotus was considered a synonym based on type culture WB 4909 (Raper & Fennell 1965), which was followed by Kozakiewicz (1989) and Hubka et al. (2013a). Aspergillus equitis was proposed as epithet for the anamorph of Eurotium chevalieri (Samson & Gams 1985). It was synonymized with A. chevalieri by Hubka et al. (2013a).
Aspergillus cibarius S.B. Hong & Samson, J. Microbiol. 50: 713. 2012. MycoBank MB800861. Fig. 24.
Fig. 24.
Aspergillus cibarius KACC 46346T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Typus: KACC 46346, holotype. Culture ex-type: KACC 46346 = DTO 197-D3 = IBT 32307.
ITS barcode: JQ918177. (Alternative markers: BenA = JQ918180; CaM = JQ918183; RPB2 = JQ918186).
Colony diam, 7 d (mm): CYA 16–18; MEA 2–10; CY20S 18–32; CY20S 30 °C 2–5; CY20S 37 °C No growth; M40Y 65–75; M60Y 65–>75; M60Y 30 °C 60–>75; M60Y 37 °C 0–9; CYAS 31–42; DG18 40–43; MEA10S 43–50.
Colony characters: CY20S 25 °C, 7 d: Colonies low to moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse straw (46). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium orange (7) at centre, sulphur yellow (15) at edge, later turn to rust (39); texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation absent or moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium orange (7) at centre, sulphur yellow (15) at edge; texture floccose; sporulation absent or sparse; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse straw (46). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium orange (7) at centre, sulphur yellow (15) at edge; texture floccose; sporulation absent or sparse; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse straw (46). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium orange (7) at centre, sulphur yellow (15) at edge; margins entire; texture floccose; sporulation absent or sparse; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse straw (46).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 100–200 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies rough along equatorial ridges, 4–5.5 × 3–5 μm, in side view lenticular, furrow present, crests irregular, < 0.5 μm. Conidiophores with smooth stipes, hyaline or light brown, 500–700 × 8–14 μm. Vesicles globose to subglobose, 32–58 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 6–11 × 3–5.5 μm. Conidia subglobose to ellipsoidal, tuberculate, 4–7 × 3.5–5.5 μm.
Distinguishing characters: Phylogenetically A. cibarius is distinct from other taxa in sect. Aspergillus, located at a basal position adjacent to the A. ruber and A. glaucus clades. Morphologically, the size, ornamentation and irregular crests of ascospores of A. cibarius resemble those of A. aurantiacoflavus, A. cumulates, A. niveoglaucus and A. xerophilus, but A. aurantiacoflavus produces orange and yellow colonies and grows slower on CYA and MEA, A. cumulatus produces globose conidia, A. niveoglaucus does not grow on CY20S at 30 °C, A. xerophilus is more xerophilic and does not grow on CYA and MEA.
Additional materials examined: China, Hebei, soil, 2001, CGMCC 3.06498 = DTO 348-H7. China, 1952, CGMCC 3.00450 = DTO 348-B5. China, tea, CGMCC 3.00451. China, Hebei, faeces, CGMCC 3.06501. Czech Republic, Prague, toenail of 56-year-old woman, 2010, isolated by P. Lysková, CCF 4098 = NRRL 62493 = DTO 354-I8. Czech Republic, Prague, toenail of 63-year-old man, 2012, isolated by P. Lysková, CCF 4235 = NRRL 62492 = DTO 354-I7. Spain, Nerja cave, near Málaga, cave sediment (entrance chambre), 2011, isolated by A. Nováková, CCF 4264 = DTO 354-I9. The Netherlands, black bean, 2012, isolated by M. Meijer, KACC 49766 = CCF 4784. The Netherlands, almond bar, 2014, isolated by T.V. Doorn, DTO 322-A6. The Netherlands, fruit pulp, 2014, isolated by T.V. Doorn, DTO 303-B8. USA, DC, Washington, chocolate glazed frosted donut, 2014, isolated by Ž. Jurjević, EMSL No. 2500. USA, NY, Elmsford, valet drover, swab, 2014, isolated by Ž. Jurjević, EMSL No. 2644. USA, CA, Danville, chocolate chip cookies, 2015, EMSL No. 2866. USA, DC, Washington, chocolate glazed frosted donut, 2014, isolated by Ž. Jurjević, EMSL No. 2499. USA, Pennsylvania, child's shoes, 2012, isolated by Ž. Jurjević, EMSL No. 1652 = CCF 5385 = DTO 355-G6. USA, Washington DC, chocolate glazed frosted donut, 2014, isolated by Ž. Jurjević, EMSL No. 2498 = CCF 5383 = DTO 355-G7. USA, California, Danville, chocolate chip cookies, 2015, isolated by Ž. Jurjević, EMSL No. 2865 = CCF 5384 = DTO 355-G8.
Aspergillus costiformis H.Z. Kong & Z.T. Qi, Acta Mycol. Sin. 14: 10. 1995. MycoBank MB363444. Fig. 25.
Fig. 25.
Aspergillus costiformis CBS 101749T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Synonyms: Eurotium costiforme H.Z. Kong & Z.T. Qi, Acta Mycol. Sin. 14: 10. 1995.
Typus: HMAS 62766, holotype. Culture ex-type: CBS 101749 = CGMCC 3.04664 = DTO 348-D8 = IBT 34456 = IBT 33662.
ITS barcode: HE615136. (Alternative markers: BenA = HE801338; CaM = HE801320; RPB2 = HE801309).
Colony diam, 7 d (mm): CYA 9–10; MEA 13–18; CY20S 40–41; CY20S 30 °C 35–42; CY20S 37 °C No growth; M40Y 60–61; M60Y 47–54; M60Y 30 °C 60–70; M60Y 37 °C 70–>75; CYAS 9–11; DG18 36–38; MEA10S 24–25.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture velvety; sporulation absent; soluble pigments absent; exudates absent; reverse sulphur yellow (15). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture velvety; sporulation absent; soluble pigments absent; exudates absent; reverse luteous (12). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture velvety; sporulation absent; soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) and white; texture velvety; sporulation absent; soluble pigments absent; exudates absent; reverse straw (46). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture velvety; sporulation absent; soluble pigments absent; exudates absent; reverse straw (46). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium sulphur yellow (15); margins entire; texture velvety; sporulation sparse; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse straw (46).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 100–255 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies rugulose, 5.5–7 × 5–6.5 μm, in side view lenticular, furrow pronounced, crests 0.5 μm. Conidiophores with smooth stipes, hyaline or light brown, 500–800 × 7–13 μm. Vesicles globose to subglobose, 20–45(–60) μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 6–9.5 × 3–4.5(–5.5) μm. Conidia globose to subglobose, microtuberculate, 4–5.5(–6.5) × 3–4.5(–5.5) μm.
Distinguishing characters: Aspergillus costiformis is characterized by large, rugulose ascospores and microtuberculate conidia. Aspergillus neocarnoyi also produces large, verruculose to rugulose ascospores, but differs in larger, tuberculate conidia measuring 8–15.5 × 6–10 μm.
Additional materials examined: China, Hebei, moldy box, 2001, CGMCC 3.06520 = DTO 348-I5. Czech Republic, Prague, toenail of 5-year-old boy, 2010, isolated by P. Lysková, CCF 4097 = NRRL 62483 = DTO 354-I3. The Netherlands, cellophane, 2015, isolated by J. Houbraken, DTO 326-B4.
Aspergillus cristatus Raper & Fennell, Gen. Aspergillus: 169. 1965. MycoBank MB326622. Fig. 26.
Fig. 26.
Aspergillus cristatus CBS 123.53T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Synonyms: Eurotium cristatum (Raper & Fennell) Malloch & Cain, Canad. J. Bot. 50: 64. 1972.
Aspergillus cristatellus Kozak., Mycol. Pap. 161: 81. 1989.
Typus: IMI 172280, neotype (Hubka et al. 2013a). Culture ex-type: CBS 123.53 = NRRL 4222 = ATCC 16468 = BCRC 33090 = FRR 1167 = IBT 5355 = IHEM 5619 = IMI 172280 = JCM 1569 = MUCL 15644 = WB 4222.
ITS barcode: EF652078. (Alternative markers: BenA = EF651914; CaM = EF652001; RPB2 = EF651957).
Colony diam, 7 d (mm): CYA 20–32; MEA 18–36; CY20S 57–75; CY20S 30 °C 55–70; CY20S 37 °C 42–51; M40Y >75; M60Y >75; M60Y 30 °C >75; M60Y 37 °C >75; CYAS 35–56; DG18 38–53; MEA10S 43–70.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium sulphur yellow (15) or orange (7); texture velvety to floccose; sporulation absent to dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse straw (46) or sulphur yellow (15) or fulvous (43). M40Y 25 °C, 7 d: Colonies moderately deep, slightly sulcate or plane; margins entire; mycelium sulphur yellow (15); texture velvety to floccose; sporulation absent to dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse to moderately dense; conidia en masse grayish green (50) to dark green (21); soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation absent to moderately dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse ochreous (44) fading into straw (46) or sulphur yellow (15). DG18 25 °C, 7 d: Colonies moderately deep, slightly sulcate or plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation absent to moderately dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse straw (46), ochreous (44) or sulphur yellow (15). MEA10S 25 °C, 7 d: Colonies moderately deep, plane or slightly sulcate; mycelium sulphur yellow (15); margins entire; texture floccose; sporulation moderately dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 100–200 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies verruculose to rugulose, 4.5–6 × 4–6 μm, in side view lenticular, furrow present, crests 1.2–1.5 μm. Conidiophores with smooth stipes, hyaline or light brown, 300–500 × (6–)8–12 μm. Vesicles globose to subglobose, (26–)35–51 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 5.5–9 × 3.5–6 μm. Conidia globose, subglobose to ellipsoidal, tuberculate, 4–6.5 × 3.5–5 μm.
Distinguishing characters: Phylogenetically A. cristatus is closely related to A. chevalieri and A. costiformis, but A. chevalieri produces smooth to slightly verruculose ascospores, and A. costiformis produces larger ascospores and microtuberculate conidia.
Additional materials examined: China, unknown source, CGMCC 3.02167, CGMCC 3.03972, CGMCC 3.06140, CGMCC 3.06141, CGMCC 3.00449, CGMCC 3.06139, CGMCC 3.00448, CGMCC 3.00463. China, Hubei, tea, CGMCC 3.07927. China, Zhejiang, tea, CGMCC 3.07934. China, Beijing, unknown source, CGMCC 3.06131. China, Yuannan, tea, CGMCC 3.07925, CGMCC 3.07926. China, Sichuan, tea, CGMCC 3.07924. China, Yunnan, Pu'er tea, CGMCC 3.15365. China, Hunan, tea, CGMCC 3.07928. China, Hunan, Fuzhuan brick tea, CGMCC 3.06086, CGMCC 3.06088, CGMCC 3.06087, CGMCC 3.06089, CGMCC 3.07930. China, Chongqing, tea, CGMCC 3.07929. China, Guangxi, tea, CGMCC 3.06083. China, Liaoning, unknown source, CGMCC 3.06085. China, Hubei, soil, CGMCC 3.06082. China, Hunan, tea, CGMCC 3.06084. China, Hunan, tea block, 2013, isolated by Q.L. Pan & L. Wang, CCF 4701 = DTO 355-B1. China, Guangxi, tea block, 2013, isolated by Q.L. Pan & L. Wang, CCF 4702 = DTO 355-B2. China, Hubei, soil, 2001, CGMCC 3.06081 = DTO 348-E9. Zaire, Kinshasa, soil, 1984, IHEM 2423 = DTO 355-B3.
Aspergillus cumulatus D.H. Kim & S.B. Hong, J. Microbiol. Biotechnol. 24: 335. 2014. MycoBank MB807118. Fig. 27.
Fig. 27.
Aspergillus cumulatus KACC 47316T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Typus: KACC 47316, holotype. Culture ex-type: KACC 47316 = DTO 303-D9 = IBT 34470 = IBT 33670.
ITS barcode: KF928303. (Alternative markers: BenA = KF928297; CaM = KF928300; RPB2 = KF928294).
Colony diam, 7 d (mm): CYA 7–10; MEA 4–5; CY20S 28–35; CY20S 30 °C 9–17; CY20S 37 °C No growth; M40Y 70–75; M60Y >75; M60Y 30 °C 62–70; M60Y 37 °C No growth; CYAS 33–42; DG18 35–47; MEA10S 65–68.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, plane to sulcate; margins irregular; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse olivaceous buff (89) at centre fading into sulphur yellow (15). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins irregular; mycelium sulphur yellow (15) and orange (7); texture floccose; sporulation moderately dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins irregular; mycelium sulphur yellow (15) and orange (7); texture floccose; sporulation moderately dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins irregular; mycelium sulphur yellow (15) and orange (7); texture floccose; sporulation moderately dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins irregular; mycelium sulphur yellow (15) and orange (7); texture floccose; sporulation moderately dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse buff (45) or sulphur yellow (15). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium sulphur yellow (15) and orange (7); margins irregular; texture floccose; sporulation moderately dense; conidia en masse dark green (21) or yellow-green (71); soluble pigments absent; exudates absent; reverse luteous (12).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 100–200 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies slightly rough, 4–6 × 3.5–5 μm, in side view lenticular, furrow pronounced, crests irregular, < 0.5 μm. Conidiophores with smooth stipes, hyaline or light brown, 500–1 300 × 7–15 μm. Vesicles globose to subglobose, 32–57 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 7–12 × 4.5–7.5 μm. Conidia globose, tuberculate, 5–8 × 4–7.5 μm.
Distinguishing characters: Phylogenetically A. cumulatus belongs to the A. ruber clade. The ascospores of A. cumulatus are unique by having irregular crests, while remaining species produce non-crested ascospores or ascospores with petaliform crests (A. appendiculatus and A. mallochii). Morphologically A. cumulatus resembles A. cibarius and A. megasporus in ascospore size and ornamentation, but A. cibarius produces subglobose to ellipsoidal conidia, A. megasporus produces larger conidia and does not grow on CY20S at 30 °C.
Additional materials examined: South Korea, air of a meju fermentation room, KACC 47513 = DTO 303-D8, KACC 47514. USA, New York, Bronx, bedroom ceiling, 2015, Ž. Jurjević, EMSL No. 2827 = CCF 5376 = DTO 355-G9.
Aspergillus endophyticus Hubka, A.J. Chen, & Samson, sp. nov. MycoBank MB818734. Fig. 28.
Fig. 28.
Aspergillus endophyticus CBS 141766T. A. Colonies: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Etymology: Name refers to its origin, isolated as endophyte of Acer pseudoplatanus.
Diagnosis: Verruculose to rugulose ascospores measuring 4–5.5 × 3–4.5 μm (crests 0.5–1 μm), tuberculate conidia measuring 5.5–8 × 4.5–6 μm.
Typus: Czech Republic, Prague, Stromovka park, endophyte of Acer pseudoplatanus, 2013, isolated by I. Kelnarová (holotype CBS H-22819, culture ex-type: CBS 141766 = DTO 354-I2 = CCF 5345 = IBT 34511).
ITS barcode: LT670941. (Alternative markers: BenA = LT671067; CaM = LT671068; RPB2 = LT671069).
Colony diam, 7 d (mm): CYA 10–12; MEA 10–12; CY20S 24–26; CY20S 30 °C No growth; CY20S 37 °C No growth; M40Y 70–75; M60Y >75; M60Y 30 °C >75; M60Y 37 °C 15–17; CYAS 30–35; DG18 35–40; MEA10S 25–35.
Colony characters: CY20S 25 °C, 7 d: Colonies low, plane; margins irregular; mycelium white to straw (46); texture floccose; sporulation sparse; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse straw (46). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium orange (7) and sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse primrose (66). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse primrose (66). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium sulphur yellow (15); margins entire; texture floccose; sporulation dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 120–200 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies verruculose to rugulose, 4–5.5 × 3–4.5 μm, in side view lenticular, furrow pronounced, crests 0.5–1 μm. Conidiophores with smooth stipes, hyaline or light brown, 350–800 × 9.5–14 μm. Vesicles globose to subglobose, 32–52 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 6–10 × 3.5–5.5 μm. Conidia globose to subglobose, tuberculate to lobate-reticulate, 5.5–8 × 4.5–6 μm.
Distinguishing characters: The phylogenetic position of A. endophyticus is not fully resolved, but it has affinity to the A. ruber and A. glaucus clades (Fig. 1). It does not grow at 30 °C and 37 °C on CY20S similarly to the majority of species from these clades, and it grows at 37 °C on M60Y similarly to A. pseudoglaucus, A. ruber, A. tonophilus and A. zutongqii (Table 5). Morphologically it resembles A. caperatus in ascospore morphology, but A. caperatus produces smaller conidia measuring 3.5–5.5 × 3.5–4.5 μm.
Aspergillus glaucus (L.) Link, Mag. Ges. Naturf. Freunde Berlin 3: 16. 1809. MycoBank MB161735. Fig. 29.
Fig. 29.
Aspergillus glaucus CBS 516.65T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Synonyms: Mucor glaucus L., Species Plantarum: 1186. 1753.
Monilia glauca (L.) Pers., Synopsis methodica fungorum: 691. 1801.
Eurotium herbariorum (Weber ex F.H. Wigg.) Link, Magazin der Gesellschaft Naturforschenden Freunde Berlin 3(1): 31. 1809.
Aspergillus herbariorum (F.H. Wigg.) E. Fisch. 1897.
Eurotium herbariorum var. minor L. Mangin, Annls Sci. Nat. Bot., Ser. 9: 365. 1909.
Aspergillus minor (L. Mangin) Thom & Raper, Department of Agriculture Miscellaneous Publications 426: 27. 1941.
Aspergillus mangini Thom & Raper, A manual of the Aspergilli: 127. 1945.
Eurotium minus (L. Mangin) Subram., Curr. Sci. 41: 760. 1972.
Aspergillus umbrosus Bainier & Sartory, Bull. Soc. Mycol. France 28 (3): 267. 1912.
Eurotium umbrosum (Bainier & Sartory) Malloch & Cain, Canad. J. Bot. 50 (1): 64. 1972.
Eurotium testaceocolorans Novobr., Novosti Sist. Nizsh. Rast. 9: 173. 1972.
Aspergillus testaceocolorans Novobr., Novosti Sist. Nizsh. Rast. 9: 173. 1972.
Typus: IMI 211383, neotype (Pitt & Samson 2000). Culture ex-type: CBS 516.65 = NRRL 116 = ATCC 16469 = DTO 197-A1 = IBT 32295 = IMI 211383 = LCP 64.1859 = Thom 5629.C = WB 116.
ITS barcode: EF652052. (Alternative markers: BenA = EF651887; CaM = EF651989; RPB2 = EF651934).
Colony diam, 7 d (mm): CYA 0–8; MEA 0–6; CY20S 25–30; CY20S 30 °C No growth; CY20S 37 °C No growth; M40Y >75; M60Y >75; M60Y 30 °C >75; M60Y 37 °C No growth; CYAS 41–49; DG18 48–60; MEA10S 60–66.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) or sulphur yellow (15) or orange (7); texture floccose; sporulation sparse; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse sulphur yellow (15) or luteous (12). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) or sulphur yellow (15) or orange (7); texture floccose; sporulation moderately dense; conidia en masse yellow-green (71) to greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) or sulphur yellow (15) or orange (7); texture floccose; sporulation sparse to moderately dense; conidia en masse pale green (19) to yellow-green (71); soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) or sulphur yellow (15) or orange (7); texture floccose; sporulation moderately dense; conidia en masse yellow-green (71); soluble pigments absent; exudates absent; reverse straw (46). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) or orange (7); texture floccose; sporulation sparse to moderately dense; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse straw (46). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium sulphur yellow (15) and white; margins entire; texture floccose; sporulation moderately dense; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse luteous (12) or ochreous (44).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 120–250 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies smooth, 5.5–7.5 × 3.5–6 μm, minute rough along equatorial ridges, in side view lenticular, furrow pronounced, crests irregular, 0.5–1 μm. Conidiophores with smooth stipes, hyaline or light brown, 150–500 × 10–21(–30) μm. Vesicles globose to subglobose, 30–60 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, (8–)12–20 × (4–)5–8.5 μm. Conidia globose, subglobose to ellipsoidal, tuberculate, 6–12.5 × 5.5–9 μm.
Distinguishing characters: Phylogenetically A. glaucus is most closely related to A. proliferans, A. aurantiacoflavus and A. niveoglaucus, but A. proliferans produces smaller (4–6 × 3–5 μm), non-crested ascospores, ascospores of A. aurantiacoflavus are also smaller (4–5.5 × 3–5 μm) and its colonies are orange-yellow. Aspergillus niveoglaucus is close to A. glaucus morphologically, but the convex surface is roughened makedly in A. niveoglaucus.
Notes: Pitt (1985) synonymized Eurotium herbariorum var. minor and A. umbrosus with A. glaucus, this was further supported by phylogenetic analyses by Hubka et al. (2013a) and our study. Aspergillus testaceocolorans was synonymized with A. glaucus by Samson, 1979, Kozakiewicz, 1989 agreed with this treatment on the basis of SEM examination, however, mistakenly synonymized A. testaceocolorans with A. pseudoglaucus (= Eurotium repens). The ex-type culture (CBS 758.74) of A. testaceocolorans is contaminated by an A. appendiculatus strain, and the position of this species could not be verified in this study.
Additional materials examined: China, unknown source, CGMCC 3.01313. China, Beijing, unknown source, CGMCC 3.06100. China, Shanxi, unknown source, CGMCC 3.06099. Puerto Rico, Bayamon, office, air, 2014, isolated by Ž. Jurjević, EMSL No. 2529 = CCF 5381 = DTO 355-H1. USA, Washington DC, unpainted board (K.B. Raper's basement), 1938, isolated by K.B. Raper, NRRL 117 = DTO 355-B4 = CCF 5582. USA, coffee beans, 1925, isolated by F.A. McCormick, NRRL 120 = 117.46 = CBS 532.65 = CCF 5583. USA, New York, Ulster Park, bedroom, settle plates, 2015, isolated by Ž. Jurjević, EMSL No. 3317 = CCF 5382 = DTO 355-H2. Unknown source, NRRL 121 = DTO 355-B5 = CCF 5584.
Aspergillus intermedius Blaser, Sydowia 28: 41. 1975. MycoBank MB309226. Fig. 30.
Fig. 30.
Aspergillus intermedius CBS 523.65T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Synonyms: Eurotium intermedium Blaser, Sydowia 28: 44. 1975.
Aspergillus spiculosus Blaser, Sydowia 28: 42. 1975.
Eurotium spiculosum Blaser, Sydowia 28: 42. 1975.
Typus: IMI 89278, neotype (Kozakiewicz 1989). Culture ex-type: CBS 523.65 = NRRL 82 = ATCC 16444 = DSM 2830 = IBT 5677 = IMI 089278ii = IMI 89278 = LSHBBB 107 = LSHTM 107 = QM 7403 = Thom 5612.107 = WB 82.
ITS barcode: EF652074. (Alternative markers: BenA = EF651892; CaM = EF652012; RPB2 = EF651958).
Colony diam, 7 d (mm): CYA 18–22; MEA 20–22; CY20S 47–55; CY20S 30 °C 45–55; CY20S 37 °C 27–36; M40Y 72–>75; M60Y 65–>75; M60Y 30 °C 65–75; M60Y 37 °C 70–>75; CYAS 29–34; DG18 39–45; MEA10S 43–54.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense or absent; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12) to sulphur yellow (15). M40Y 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense or absent; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12). M60Y 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense or absent; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense or absent; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse ochreous (44) at centre, luteous (12) to sulphur yellow (15) at edge. DG18 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense or absent; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse ochreous (44) to luteous (12) to sulphur yellow (15). MEA10S 25 °C, 7 d: Colonies moderately deep, slightly sulcate; mycelium sulphur yellow (15); margins entire; texture floccose; sporulation moderately dense or absent; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse ochreous (44) to luteous (12).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 100–250 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies verruculose to rugulose, 3.5–5 × 3–4.5 μm, in side view lenticular, furrow present, crests 0.5 μm. Conidiophores with smooth stipes, hyaline or light brown, 250–600 × 7.5–13 μm. Vesicles globose to subglobose, (26–)40–60 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 5.5–7.5(–9) × 3–5.5 μm. Conidia globose to subglobose, microtuberculate, 3–4(–6) × 3–4.5 μm.
Distinguishing characters: Phylogenetically and morphologically A. intermedius resembles A. montevidensis, A. porosus and A. caperatus, but can be distinguished by smooth conidia (microtuberculate under SEM) instead of roughened conidia (lobate-reticulate under SEM) in the other species.
Additional materials examined: China, unknown source, 1969, CGMCC 3.03968 = DTO 348-D6. China, unknown source, CGMCC 3.06138. China, Beijing, unknown source, CGMCC 3.01300. China, industrial material, 1955, isolated by V. Zánová, CCF 127 = DTO 354-I5. Croatia, unknown source, isolated by V. Johanides, CBS 108.55. Czech Republic, unknown source, 1956, CGMCC 3.00664 = DTO 348-C1. Czech Republic, Prague, sputum of 55-year-old woman, 2013, isolated by P. Lysková, CCF 4681 = DTO 354-I6. Czech Republic, Prague, air sampler, surgical operating room, 2014, isolated by A. Ešnerová, CCF 5377 = DTO 355-G5. Spain, Badajoz, soil, isolated by P. Blaser, CBS 377.75. The Netherlands, fruit jam, 2014, isolated by T. van Doorn, DTO 345-H5. USA, IL, Peoria, soy protein, isolated by A.J. Moyer, NRRL 25823. Unknown source, NRRL 84. Unknown country, butter, NRRL 4817 = DTO 355-B9 = IFO 5322 = IMI 313754 = JCM 23051 = CCF 5608.
Aspergillus leucocarpus Hadlok & Stolk, Antonie van Leeuwenhoek 35: 9. 1969. MycoBank MB326642. Fig. 31.
Fig. 31.
Aspergillus leucocarpus CBS 353.68T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Synonyms: Eurotium leucocarpum Hadlok & Stolk, Antonie van Leeuwenhoek 35: 9. 1969.
Typus: CBS 353.68, holotype. Culture ex-type: CBS 353.68 = IBT 5350 = IMI 278375 = NRRL 3497 = PIL 620 = QM 9365 = QM 9707.
ITS barcode: EF652087. (Alternative markers: BenA = EF651925; CaM = EF652023; RPB2 = EF651972).
Colony diam, 7 d (mm): CYA 24–31; MEA 21–31; CY20S 68–70; CY20S 30 °C 42–70; CY20S 37 °C No growth; M40Y >75; M60Y >75; M60Y 30 °C >75; M60Y 37 °C 35–58; CYAS 32–40; DG18 43–52; MEA10S 47–50.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) and white; texture velvety to floccose; sporulation sparse; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse straw (46). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) and white; texture floccose; sporulation sparse; conidia en masse dark green (21) to greenish olivaceous (90); soluble pigments absent; exudates absent; reverse luteous (12). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) and white; texture floccose; sporulation sparse; conidia en masse dark green (21) to greenish olivaceous (90); soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) and white; texture floccose; sporulation sparse; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse straw (46). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) and white; texture floccose; sporulation sparse; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse straw (46). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium straw (46) and white; margins entire; texture floccose; sporulation sparse; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, white, globose to subglobose, 80–140 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies verruculose, 4.5–5.5 × 3.5–5 μm, in side view lenticular, furrow present, crests 0.8–1.5 μm. Conidiophores with smooth stipes, hyaline or light brown, 800–1 400 × 7.5–12 μm. Vesicles globose to subglobose, 35–60 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 8–11.5 × 3.5–6.5 μm. Conidia globose to subglobose, tuberculate, 5.5–9 × 5–8 μm.
Distinguishing characters: The white ascomata are consistently produced in all available A. leucocarpus strains, which can easily distinguish it from other sect. Aspergillus species.
Additional materials examined: Canada, house dust, 2015, isolated by C.M. Visagie, DTO 357-A2 = KAS7576. Madagascar, vanilla sticks, 2012, isolated by J. Houbraken, DTO 174-I5.
Aspergillus levisporus Hubka, A.J. Chen, Jurjević & Samson, sp. nov. MycoBank MB818735. Fig. 32.
Fig. 32.
Aspergillus levisporus CBS 141767T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Etymology: Name refers to its smooth ascospores.
Diagnosis: Smooth ascospores measuring 3–4.5 × 2.5–4 μm.
Typus: USA, MO, Saint Louis, bedroom, wood base, 2015, isolated by Ž. Jurjević (holotype: CBS H-22820, culture ex-type: CBS 141767 = DTO 355-G4 = EMSL No.3211 = CCF 5378 = IBT 34512).
ITS barcode: LT670950. (Alternative markers: BenA = LT671094; CaM = LT671095; RPB2 = LT671096).
Colony diam, 7 d (mm): CYA 13–17; MEA 8–10; CY20S 19–20; CY20S 30 °C 18–20; CY20S 37 °C No growth; M40Y 60–65; M60Y 65–67; M60Y 30 °C 70–75; M60Y 37 °C No growth; CYAS 35–37; DG18 35–36; MEA10S 40–41.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, plane; margins irregular; mycelium sulphur yellow (15); texture floccose; sporulation dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse greenish olivaceous (90) at centre, sulphur yellow (15) at edge. M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins irregular; mycelium white; texture floccose; sporulation dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse ochreous (44). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins slightly irregular; mycelium white; texture floccose; sporulation dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse ochreous (44). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse greenish olivaceous (90) at centre, fading into olivaceous buff (89). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse sulphur yellow (15) at centre, fading into yellow-green (71). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium white; margins entire; texture floccose; sporulation moderately dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse fulvous (43) at centre, ochreous (44) at edge.
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 70–130 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies smooth, 3–4.5 × 2.5–4 μm, in side view lenticular, furrow present, crests absent. Conidiophores with smooth stipes, hyaline or light brown, 400–600 × 10–14 μm. Vesicles globose to subglobose, 30–44 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 6–8.5 × 3.5–6 μm. Conidia globose, tuberculate to lobate-reticulate, 3.5–4.5 × 2.5–4 μm.
Distinguishing characters: The non-crested, smooth ascospores of A. levisporus resemble those of A. proliferans, A. pseudoglaucus, A. ruber and A. sloanii, but the latters all produce larger ascospores, 4–6 × 3–5 μm in A. proliferans, 4–6.5 × 3–4.5 μm in A. pseudoglaucus, 4–6 × 3.5–5 μm in A. ruber and 4–6 × 3–4.5 μm in A. sloanii, respectively.
Aspergillus mallochii Visagie, Yilmaz & Seifert, MycoKeys 19: 16. 2017. MycoBank MB819025. Fig. 33.
Fig. 33.
Aspergillus mallochii CBS 141928T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Typus: DAOM 740296, holotype. Culture ex-type: CBS 141928 = DTO 357-A5 = KAS7618 = DAOMC 146054.
ITS barcode: KX450907. (Alternative markers: BenA = KX450889; CaM = KX450902; RPB2 = KX450894).
Colony diam, 7 d (mm): CYA 7–8; MEA 2–3; CY20S 11–12; CY20S 30 °C No growth; CY20S 37 °C No growth; M40Y 53–55; M60Y 64–70; M60Y 30 °C 42–47; M60Y 37 °C No growth; CYAS 29–30; DG18 35–38; MEA10S 33–35.
Colony characters: CY20S 25 °C, 7 d: Colonies low, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse sulphur yellow (15). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15), later turn into vinaceous (57) to orange (7); texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse ochreous (44). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse straw (46). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse straw (46). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium sulphur yellow (15); margins entire; texture floccose; sporulation sparse; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 130–220 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies smooth, minute rough along equatorial ridges, 4–6 × 3–5 μm, in side view lenticular, furrow absent or showing as a trace, crests petaliform, 1–2 μm at high part. Conidiophores with smooth stipes, hyaline or light brown, 600–1 500 × 6–9.5(–12) μm. Vesicles globose to subglobose, 27–43 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 6.5–9 × 3–5 μm. Conidia subglobose to ellipsoidal, tuberculate, 4.5–7 × 4–5.5 μm.
Distinguishing characters: Phylogenetically and morphologically A. mallochii is close to A. appendiculatus, but A. appendiculatus produces larger ascospores (5–7.5 × 4–5.5 μm) and does not grow on MEA and CYA at 25 °C.
Additional materials examined: The Netherlands, chocolat miroir, 2015, CBS 141776 = DTO 343-G3.
Aspergillus megasporus, Visagie, Yilmaz & Seifert, MycoKeys 19: 17. 2017. MycoBank MB819028. Fig. 34.
Fig. 34.
Aspergillus megasporus CBS 141929T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Typus: DAOM 741781, holotype. Culture ex-type: CBS 141929 = DTO 356-H7 = KAS 6176 = DAOMC 250799.
ITS barcode: KX450910. (Alternative markers: BenA = KX450892; CaM = KX450905; RPB2 = KX450897).
Colony diam, 7 d (mm): CYA 10–11; MEA 4–6; CY20S 38–40; CY20S 30 °C No growth; CY20S 37 °C No growth; M40Y 55–60; M60Y 70–75; M60Y 30 °C 61–64; M60Y 37 °C No growth; CYAS 23–24; DG18 38–40; MEA10S 50–52.
Colony characters: CY20S 25 °C, 7 d: Colonies low, plane; margins entire; mycelium straw (46) to sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse ochreous (44). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation dense; conidia en masse greyish green (50) to dark green (21); soluble pigments absent; exudates absent; reverse luteous (12). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies deep, plane; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse straw (46). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12) at centre, fading into yellow-green (71). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium sulphur yellow (15) and white; margins entire; texture floccose; sporulation moderately dense; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse ochreous (44).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 110–300 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies smooth, rough along equatorial ridges, 4–6.5 × 3.5–5.5 μm, in side view lenticular, furrow present, crests absent or indefinite. Conidiophores with smooth stipes, hyaline or light brown, 1 000–1 500 × 6.5–12(–21.5) μm. Vesicles globose to subglobose, 30–54 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 7.5–14 × 4–7.5 μm. Conidia subglobose to ellipsoidal, tuberculate, 7–14 × 5–8.5 μm.
Distinguishing characters: Aspergillus megasporus belongs to A. glaucus clade (Fig. 1). Its ascospore dimensions are similar to those of A. aurantiacoflavus, A. glaucus, A. niveoglaucus and A. proliferans. However, A. aurantiacoflavus, A. glaucus and A. niveoglaucus have low, irregular crests in contrast to non-crested ascospores in A. megasporus and A. proliferans. Aspergillus proliferans can be differentiated by smaller conidia.
Additional materials examined: Canada, New Brunswick, Little Lepreau, house dust, 2015, isolated by C.M. Visagie, DTO 356-H1 = KAS5973 = DAOMC 250800. The Netherlands, Dutch chocolate butter, 2007, isolated by M. Meijer, CBS 141772 = DTO 048-I3.
Aspergillus montevidensis Talice & Mackinnon, Compt. Rend. Soc. Biol. Fr. 108: 1007. 1931. MycoBank MB309231. Fig. 35.
Fig. 35.
Aspergillus montevidensis CBS 491.65T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Synonyms: Eurotium montevidense (Talice & J.A. Mackinnon) Malloch & Cain, Canad. J. Bot. 50 (1): 64. 1972.
Eurotium amstelodami var. montevidense (Talice & J.A. Mackinnon) Kozak., Mycol. Pap. 161: 86. 1989.
Aspergillus vitis var. montevidensis Kozak., Mycol. Pap. 161: 86. 1989.
Aspergillus heterocaryoticus C.M. Chr., L.C. López & C.R. Benj., Mycologia 57 (4): 535. 1965.
Eurotium heterocaryoticum C.M. Chr., L.C. López & C.R. Benj., Mycologia 57 (4): 536. 1965.
Aspergillus vitis Novobr., Novosti Sist. Nizsh. Rast. 9: 175. 1972.
Eurotium vitis Novobr., Novosti Sist. Nizsh. Rast. 9: 175. 1972.
Aspergillus hollandicus Samson & W. Gams, Advances in Penicillium and Aspergillus Systematics: 33. 1985.
Typus: BPI 884202, neotype (Hubka et al. 2013a). Culture ex-type: CBS 491.65 = NRRL 108 = ATCC 10077 = IBT 5685 = IHEM 3337 = IMI 172290 = NRRL 109 = QM 7423 = Thom 5290 = Thom 5633.24 = WB 108.
ITS barcode: EF652077. (Alternative markers: BenA = EF651898; CaM = EF652020; RPB2 = EF651964).
Colony diam, 7 d (mm): CYA 19–24; MEA 18–23; CY20S 45–61; CY20S 30 °C 25–50; CY20S 37 °C 28–30; M40Y 60–>75; M60Y 60–>75; M60Y 30 °C 60–>75; M60Y 37 °C >75; CYAS 20–47; DG18 38–60; MEA10S 53–65.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium sulphur yellow (15) and white; texture velvety to floccose; sporulation sparse to moderately dense; conidia en masse greyish green (50) or white; soluble pigments absent; exudates absent; reverse greenish olivaceous (90) or sulphur yellow (15). M40Y 25 °C, 7 d: Colonies moderately deep, slightly sulcate or plane; margins entire; mycelium sulphur yellow (15) and white; texture velvety to floccose; sporulation sparse to moderately dense; conidia en masse greyish green (50) or white; soluble pigments absent; exudates absent; reverse ochreous (44) or luteous (12). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) and white; texture velvety or floccose; sporulation sparse to moderately dense; conidia en masse greyish green (50) or white; soluble pigments absent; exudates absent; reverse ochreous (44) or luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, plane or sulcate; margins slightly irregular; mycelium white and sulphur yellow (15); texture floccose; sporulation sparse to moderately dense; conidia en masse greyish green (50) or glaucous (73) or white; soluble pigments absent; exudates absent; reverse greenish olivaceous (90) to salmon (41). DG18 25 °C, 7 d: Colonies moderately deep, plane to slightly sulcate; margins entire; mycelium sulphur yellow (15) and white; texture velvety to floccose; sporulation moderately dense to dense; conidia en masse greyish green (50) or white; soluble pigments absent; exudates absent; reverse greenish olivaceous (90) to sulphur yellow (15). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium sulphur yellow (15) and white; margins entire; texture velvety to floccose; sporulation moderately dense or absent; conidia en masse greyish green (50) or white; soluble pigments absent; exudates absent; reverse luteous (12) to ochreous (44).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 80–250 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies in most strains rugulose; smooth or slightly rough in atypical strain CCF 4070, tuberculate in atypical strain CCF 4248, 4–6 × 3–4.5 μm, in side view lenticular, furrow pronounced, crests 0.5 μm. Conidiophores with smooth stipes, hyaline or light brown, 250–500 × 6–13.5 μm. Vesicles globose to subglobose, 25–35(–50) μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 5–8.5(–11) × 3–6 μm. Conidia globose, subglobose to ellipsoidal, lobate-reticulate, 4–6.5 × 3.5–5 μm.
Distinguishing characters: Morphologically and phylogenetically A. montevidensis is close to A. intermedius, but A. intermedius produces microtuberculate conidia.
Notes: The recent species concept of A. amstelodami sensu (Thom and Raper, 1941, Raper and Fennell, 1965, Blaser, 1975) is different from the original description (Mangin 1909). Pitt (1985) speculated that the original strain had been replaced by the species described in Thom & Raper (1941), and recommended A. montevidensis as a substitute name for A. amstelodami. Hubka et al. (2013a) agreed and considered the description of A. montevidensis (Talice & Mackinnon 1931) the first valid description of the species consistent with E. amstelodami sensu Thom & Raper (1941). Aspergillus hollandicus and A. vitis were proposed for the anamorphic name of E. amstelodami Mangin (Samson & Gams 1985, Kozakiewicz 1989). Aspergillus heterocaryoticus was considered to be conspecific with E. amstelodami (Blaser, 1975, Samson, 1979). These three species were synonymized with A. montevidensis (Hubka et al. 2013a).
Additional materials examined: China, mite, 1969, CGMCC 3.03888 = DTO 348-D3. China, Ningxia, unknown source, CGMCC 3.06069, CGMCC 3.06072. China, unknown source, CGMCC 3.00462, CGMCC 3.01307, CGMCC 3.00771, CGMCC 3.01306, CGMCC 3.01309, CGMCC 3.04462, CGMCC 3.06064, CGMCC 3.03967, CGMCC 3.04059, CGMCC 3.01304, CGMCC 3.01308. China, Hebei, unknown source, CGMCC 3.06074. China, Neimenggu, unknown source, CGMCC 3.06071, CGMCC 3.06077, CGMCC 3.06078, CGMCC 3.06073. China, Hebei, soil, CGMCC 3.06511. China, Henan, corn, CGMCC 3.06065. China, Ningxia, soil, CGMCC 3.06066. China, Hebei, moldy agaric, CGMCC 3.06513. China, Yunnan, moldy bean curd, CGMCC 3.06517. China, Hebei, straw, CGMCC 3.06512. China, moldy sugarcane, CGMCC 3.07157. China, Hebei, moldy pine seeds, CGMCC 3.06514. China, Xinjiang, soil, CGMCC 3.11413. China, Beijing, unknown source, CGMCC 3.06063, CGMCC 3.06075, CGMCC 3.06076. China, Hainan, soil, CGMCC 3.06061. China, Guizhou, soil, CGMCC 3.06068. China, Ningxia, soil, CGMCC 3.06070. China, Jiangsu, fabric, CGMCC 3.07178. China, Xinjiang, soil, CGMCC 3.11525, CGMCC 3.11410. China, Hebei, soil, CGMCC 3.06510. China, Hebei, moldy bark, CGMCC 3.06516. China, Yunnan, moldy bamboo, CGMCC 3.06518. China, Hunan, soil, CGMCC 3.06067. China, Hebei, moldy leaves, CGMCC 3.06515. China, industrial material, 1955, isolated by V. Zánová, CCF 726. Czech Republic, feed, 1984, isolated by V. Neumannová, CCF 1952. Czech Republic, Prague, neck skin of 78-year-old woman, 2008, isolated by M. Skořepová, CCF 3998. Czech Republic, heel skin of 32-year-old man, Prague, 2007, isolated by M. Skořepová, CCF 4069. Czech Republic, fingernail of 32-year-old woman, Prague, 2007, isolated by M. Skořepová, CCF 4070. Czech Republic, Prague, thigh and neck skin of 42-year-old woman, 2010, isolated by P. Lysková, CCF 4071. Czech Republic, Skrbeň, window sill, 1997, isolated by A. Kubátová, CCF 4248. Czech Republic, České Budějovice, sputum of 11-year-old girl, 2010, isolated by N. Mallátová, CCF 4258. Czech Republic, Prague, bronchoalveolar lavage of 40-year-old man, 2012, isolated by P. Lysková, CCF 4370. Czech Republic, Prague, external auditory canal of 66-year-old man, 2010, isolated by P. Lysková, CCF 4371. Czech Republic, Prague, bronchoalveolar lavage fluid of 60-year-old male, 2015, isolated by P. Lysková, PL 378/15. Czech Republic, Prague, air sampler – intensive care unit room (haematooncology), 2013, isolated by V. Chrenková, MY1832. Czech Republic, Prague, air sampler – paediatric haematooncology unit, 2013, isolated by V. Chrenková, MY2467. Czech Republic, Prague, lungs of 43-year-old woman after lung transplantation, 2014, isolated by V. Chrenková, MY4449. Czech Republic, Prague, fingernail of 37-year-old man, 2007, isolated by M. Skořepová, SK237. Czech Republic, Prague, pigeon dropping, 1991, isolated by K. Prášil and R. Kolínská, CCF 2723. Czech Republic, Prague, white Arabic bread (pita), 1999, isolated by A. Kubátová, CCF 3750. Czech Republic, Veleliby near Nymburk, seeds of Papaver somniferum, 1999, isolated by J. Hubert, CCF 3135. Denmark, straw, 2012, isolated by J. Houbraken, DTO 212-D3. Germany, bakery, 2010, isolated by T. Hoogenhuijzen, DTO 121-G7. Hungary, table, 2009, isolated by van Mil, DTO 101-F5. Hungary, indoor air, 2014, isolated by M. Meijer, DTO 147-I4. Kazakhstan, Alma-Ata, ex grapes, 1968, isolated by L.A. Beljakova, CBS 651.74 = ATCC 24717 = IMI 174724 = VKM F-1760. Mexico, Oryza sativa kernel, 1963, isolated by C.R. Benjamin, NRRL A-13891 = CBS 410.65. Spain, Cantabria, Altamira Cave, cave sediment, 2008, isolated by A. Nováková, S14. Suriname, plywood, isolated by M.B. Schol-Schwarz, CBS 111.52 = DTO 351-C9. The Netherlands, cake, 2015, isolated by M. Meijer, DTO 334-A3. The Netherlands, corn kernels (imported), 2014, isolated by J. Houbraken, DTO 300-E3. The Netherlands, sesame seed (imported), 2013, isolated by J. Houbraken, DTO 253-H7. USA, unknown source, ∼1910, NRRL 90 = CBS 518.65. USA, Missouri, Columbia, candied grapefruit rind, isolated by D.I. Fennell, NRRL 4716. USA, IL, Peoria, refrigerated bread dough, isolated by R. Graves, NRRL 25850. USA, IL, Chicago, nasal swab, NRRL 35697. USA, PA, Mahanoy City, bedroom, settle plates, 2015, isolated by Ž. Jurjević, EMSL No. 2934 = CCF 5379 = DTO 355-H3. USA, Delaware, green house, air, 2011, isolated by Ž. Jurjević, EMSL No. 1589. USA, FL, Loxahatchee, Home, Kitchen cabinet, 2013, isolated by Ž. Jurjević, EMSL No. 2187. USA, IL, Chicago, bathroom, settle plates, 2015, isolated by Ž. Jurjević, EMSL No. 2790. Unknown source, NRRL 89.
Aspergillus neocarnoyi Kozak., Mycol. Pap. 161: 63. 1989. MycoBank MB127756. Fig. 36.
Fig. 36.
Aspergillus neocarnoyi CBS 471.65T. A. Colonies: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Synonyms: Aspergillus carnoyi (Biourge) Thom & Raper, Misc. Publ. U.S. Dept. Agric.: 34. 1941, nom. inval. [Art. 39.1 McNeill et al. 2012]
Eurotium carnoyi Malloch & Cain, Canad. J. Bot. 50 (1): 63. 1972.
Typus: IMI 172279, holotype. Culture ex-type: CBS 471.65 = NRRL 126 = ATCC 16924 = IBT 6016 = IMI 172279 = LSHTM A32 = QM 7402 = Thom 5612.A32 = WB 126 = DTO 196-H6.
ITS barcode: EF652057. (Alternative markers: BenA = EF651903; CaM = EF651985; RPB2 = EF651942).
Colony diam, 7 d (mm): CYA No growth; MEA No growth; CY20S 3–5; CY20S 30 °C No growth; CY20S 37 °C No growth; M40Y 20–25; M60Y 53–65; M60Y 30 °C 15–18; M60Y 37 °C No growth; CYAS 18–20; DG18 32–42; MEA10S 35–38.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) and white; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse straw (46). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium orange (7) at centre, white at edge; texture floccose; sporulation sparse; conidia en masse green (20); soluble pigments absent; exudates absent; reverse luteous (12) fading into saffron (10). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium orange (7) at centre, white at edge; texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44) fading into straw (46). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium fulvous (43) at centre, white at edge; texture floccose; sporulation sparse; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12) fading into straw (46). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium saffron (10) at centre, white at edge; margins entire; texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse rust (39) at centre, fading into saffron (10).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 120–230 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies verruculose to rugulose, 6.5–9 × 4.5–7 μm, in side view lenticular, furrow present, crests absent or indefinite. Conidiophores with smooth stipes, hyaline or light brown, 1 000–2 000 × (9–)12–23 μm. Vesicles globose to subglobose, (32–)50–92 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 12–21 × 6–9 μm. Conidia ellipsoidal, tuberculate, 8–15.5 × 6–10 μm.
Distinguishing characters: Phylogenetically A. neocarnoyi is closely related to A. brunneus and A. niveoglaucus, but A. brunneus produces mainly globose conidia, while A. niveoglaucus produces smaller ascospores. The large ascospores of A. neocarnoyi also resemble those of A. osmophilus, but A. osmophilus produces smaller conidia and ascospores with thick crests.
Additional materials examined: Slovenia, Ljubljana, Slovene Ethnographic museum, air at the sampling of shaman statue originating from Mali, 2016, isolated by P. Zalar, EXF-10029 = DTO 357-E2.
Aspergillus niveoglaucus Thom & Raper, U.S.D.A. Misc. Pub. 426: 35. 1941. MycoBank MB120985. Fig. 37.
Fig. 37.
Aspergillus niveoglaucus CBS 114.27T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Synonyms: Eurotium niveoglaucum (Thom & Raper) Malloch & Cain, Canad. J. Bot. 50 (1): 64. 1972.
Aspergillus glauconiveus Samson & W. Gams, Advances in Penicillium and Aspergillus Systematics: 45. 1985.
Aspergillus parviverruculosus H.Z. Kong & Z.T. Qi, Acta Mycol. Sin. 14 (1): 12. 1995.
Eurotium parviverruculosum H.Z. Kong & Z.T. Qi, Acta Mycol. Sin. 14 (1): 12. 1995.
Typus: IMI 32050ii, neotype (Samson & Gams 1985). Culture ex-type: CBS 114.27 = CBS 517.65 = NRRL 127 = ATCC 10075 = BCRC 33096 = CGMCC 3.4374 = FRR 927 = IBT 5356 = IMI 32050 = JCM 1578 = LSHBA 16 = NRRL 129 = NRRL 130 = QM 1977 = Thom 5612.A16 = Thom 5633 = Thom 5633.7 = Thom 7053.2 = UAMH 6591 = WB 127 = WB 130.
ITS barcode: EF652058. (Alternative markers: BenA = EF651905; CaM = EF651993; RPB2 = EF651943).
Colony diam, 7 d (mm): CYA 2–8; MEA 0–5; CY20S 12–30; CY20S 30 °C No growth; CY20S 37 °C No growth; M40Y >75; M60Y >75; M60Y 30 °C 65–>75; M60Y 37 °C No growth; CYAS 32–37; DG18 34–42; MEA10S 40–45.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) or white; texture floccose; sporulation sparse; conidia en masse white or pale green (19); soluble pigments absent; exudates absent; reverse straw (46). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium rosy buff (61) or straw (46) or white; texture floccose; sporulation sparse; conidia en masse pale green (19) or white; soluble pigments absent; exudates absent; reverse apricot (42). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) or white; texture floccose; sporulation sparse; conidia en masse white or pale green (19); soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) or white; texture floccose; sporulation moderately dense; conidia en masse white or pale green (19); soluble pigments absent; exudates absent; reverse straw (46). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) or white; texture floccose; sporulation moderately dense; conidia en masse white or pale green (19); soluble pigments absent; exudates absent; reverse straw (46). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium straw (46) or white; margins entire; texture floccose; sporulation moderately dense; conidia en masse white or pale green (19); soluble pigments absent; exudates absent; reverse luteous (12).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 90–240 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies rough along equatorial ridges or verruculose to rugulose, (4.5–)5.5–7.5 × (3–)5–6 μm, in side view lenticular, furrow present, crests irregular, < 0.5 μm. Conidiophores with smooth stipes, hyaline or light brown, 1 000–1 500 × (7.5)–10–23 μm. Vesicles globose to subglobose, (31–)55–85 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 8–14(–20) × 4–7(–11) μm. Conidia subglobose to ellipsoidal, tuberculate, (6–)8–13.5 × 4–9 μm.
Distinguishing characters: Phylogenetically A. niveoglaucus is closely related to A. brunneus and A. neocarnoyi, but these two species produce larger ascospores, 7–10 × 6–8 μm in A. brunneus and 6.5–9 × 4.5–7 μm in A. neocarnoyi. Morphologically, it resembles A. glaucus in ascospore size and ornamentation, but the convex surface is less roughened in ascospores of A. glaucus.
Additional materials examined: Belgium, Namur, indoor air, 1983, IHEM 1811 = DTO 355-C3. Brazil, corn kernels, 2008, isolated by J. Houbraken, DTO 060-I3. Canada, Manitoba, Barley feed, isolated by M. Desjardins, CBS 117311. China, Hebei, soil, CBS 101750 = CGMCC 3.04665 (AS 3.4665) = DTO 197-B4. China, Guangdong, cashew Kernel, 2001, CGMCC 3.06092 = DTO 348-F3. China, Yunnan, moldy corn leaves, CGMCC 3.06496. China, Neimenggu, soil, CGMCC 3.07854. China, Guizhou, unknown source, CGMCC 3.06102. China, unknown source, CGMCC 3.01312, CGMCC 3.01294. Czech Republic, garlic, isolated by L. Marvanová, CCM F-530 = CCF 4038. Czech Republic, Prague, cereals, 1993, isolated by A. Kubátová, CCF 4388. South Korea, soybeans, 2012, isolated by D.H. Kim, CCF 4787 = KACC 47144 = DTO 355-C4, CCF 4790 = KACC 47147 = DTO 355-C5. Spain, Andalusia, Málaga, Cueva del Tesoro, cave sediment from the cave wall, 2010, A. Nováková, CCF 4191 = DTO 355-C1. The Netherlands, apricot paste, 2014, isolated by M. Meijer, DTO 308-B9. The Netherlands, animal feed kernels, 2016, isolated by J. Dijksterhuis, DTO 346-B4. The Netherlands, spoiled starch, isolated by J. Houbraken, DTO 193-B6. USA, Montana, Great Falls, air of bathroom, 2013, isolated by Ž. Jurjević, EMSL No. 2211 = CCF 5380 = DTO 355-H8. Unknown source, isolated by G. Smith, NRRL 128, NRRL 136, NRRL 137.
Aspergillus osmophilus Asgari & Zare, Mycoscience 55: 58. 2013. MycoBank MB803278. Fig. 38.
Fig. 38.
Aspergillus osmophilus CBS 134258T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Typus: IRAN 16110 F, holotype. Culture ex-type: CBS 134258 = IRAN 2090C = DTO 354-C1.
ITS barcode: KC473921. (Alternative markers: BenA = LT671127; CaM = LT671128; RPB2 = LT671129).
Colony diam, 7 d (mm): CYA No growth; MEA No growth; CY20S No growth; CY20S 30 °C 2–3; CY20S 37 °C No growth; M40Y 30–41; M60Y >75; M60Y 30 °C >75; M60Y 37 °C 63–65; CYAS 6–7; DG18 43–45; MEA10S 54–60.
Colony characters: M40Y 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium salmon (41); texture floccose; sporulation sparse; conidia en masse pale green (19); soluble pigments absent; exudates light yellow droplets; reverse fulvous (43). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium primrose (66); texture floccose; sporulation sparse; conidia en masse pale green (19); soluble pigments absent; exudates light yellow droplets; reverse ochreous (44). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium primrose (66); texture floccose; sporulation absent; soluble pigments light brown; exudates absent; reverse orange (7) at centre, saffron (10) at edge. DG18 25 °C, 7 d: Colonies moderately deep, plane; margins irregular; mycelium salmon (41); texture floccose; sporulation sparse; conidia en masse pale green (19); soluble pigments absent; exudates light yellow droplets; reverse fulvous (43). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium primrose (66); margins entire; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse ochreous (44).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 100–350 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies verruculose, 7–9 × 6–7.5 μm, in side view lenticular, furrow pronounced, crests 0.5 μm. Conidiophores with smooth stipes, hyaline or light brown, 300–1 000 × 7.5–12 μm. Vesicles globose to subglobose, 28–46 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 9–12 × 4.5–7 μm. Conidia subglobose to ellipsoidal, microtuberculate to tuberculate, 6–8.5 × 5.5–7.5 μm.
Distinguishing characters: Phylogenetically A. osmophilus is closely related to A. xerophilus, but A. xerophilus produces smaller ascospores (4.5–6.5 × 3.5–5 μm) and does not grow on M60Y at 37 °C. The large ascospores of A. osmophilus resemble those of A. brunneus and A. neocarnoyi, but A. brunneus can grow on CYA and MEA, A. neocarnoyi produces larger conidia measuring 8–15.5 × 6–10 μm.
Aspergillus porosus A.J. Chen, Frisvad & Samson, sp. nov. MycoBank MB818736. Fig. 39.
Fig. 39.
Aspergillus porosus CBS 141770T. A. Colonies: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Etymology: Name refers to small holes on the ascospores.
Diagnosis: Small, pitted ascospores (3.5–5.5 × 3–4.5 μm), lobate-reticulate conidia (3.5–5.5 × 2.5–4.5 μm).
Typus: Turkey, soil, 2013, isolated by Canan Unal (holotype: CBS H-22822, culture ex-type: CBS 141770 = DTO 262-D7 = IBT 34443).
ITS barcode: LT670961. (Alternative markers: BenA = LT671130; CaM = LT671131; RPB2 = LT671132).
Colony diam, 7 d (mm): CYA 21–23; MEA 18–19; CY20S 58–60; CY20S 30 °C 37–58; CY20S 37 °C 31–33; M40Y >75; M60Y >75; M60Y 30 °C >75; M60Y 37 °C >75; CYAS 35–41; DG18 45–50; MEA10S 62–63.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium sulphur yellow (15); texture velvety; sporulation moderately dense or absent; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12) to ochreous (44). M40Y 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12). M60Y 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation absent to sparse; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse ochreous (44) fading into sulphur yellow (15). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse to moderately dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12) to sulphur yellow (15). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium sulphur yellow (15); margins entire; texture floccose; sporulation sparse to moderately dense; conidia en masse dark green (21); soluble pigments absent; exudates absent; reverse luteous (12) to ochreous (44).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 80–230 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies rugulose, pitted, 3.5–5.5 × 3–4.5 μm, in side view lenticular, furrow pronounced, crests 0.5 μm. Conidiophores with smooth stipes, hyaline or light brown, 250–600 × 5–12.5 μm. Vesicles globose to subglobose, 24–58 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 5–10 × 2.5–5 μm. Conidia globose to subglobose, lobate-reticulate, 3.5–5.5 × 2.5–4.5 μm.
Distinguishing characters: Under SEM, the entire surface of ascospores of A. porosus is pitted, in contrast most sect. Aspergillus species have holes along equatorial ridges. Phylogenetically A. porosus is related to A. caperatus, A. intermedius and A. montevidensis, but A. intermedius can be distinguished by smooth conidia (microtuberculate under SEM), A. caperatus does not grow on CY20S at 37 °C, and A. montevidensis produces slightly larger conidia (4–6.5 × 3.5–5 μm).
Additional materials examined: Israel, Arachis hypogaea fruit, isolated by P. Blaser, CBS 375.75 = DTO 197-C4. South Africa, Robben Island, soil, 2015, isolated by M. Meijer, DTO 338-A7. Turkey, soil, 2014, isolated by R. Demirel, DTO 308-D1. Turkey, soil, 2013, isolated by A. Yoltas, DTO 262-D4, DTO 262-D2.
Aspergillus proliferans G. Sm., Trans. Brit. Mycol. Soc. 26: 26. 1943. MycoBank MB284312. Fig. 40, Fig. 41.
Fig. 40.
Aspergillus proliferans CBS 121.45T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, C, F. Conidiophores. D, G. Conidia. E. Ascomata initials. Scale bars: B = 20 μm; C–E = 10 μm; F = 20 μm; G = 2 μm.
Fig. 41.
Aspergillus proliferans DTO 322-A2. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Synonyms: Aspergillus acutus Blaser, Sydowia 28: 33. 1975.
Eurotium acutum Blaser, Sydowia 28: 33. 1975.
Typus: IMI 016105iii, lectotype (Samson & Gams 1985). Culture ex-type: CBS 121.45 = NRRL 1908 = IBT 6213 = IMI 016105ii = IMI 016105iii = IMI 016105 = LSHB BB.82 = MUCL 15625 = NCTC 6546 = QM 7462 = UC 4303 = WB 1908.
ITS barcode: EF652064. (Alternative markers: BenA = EF651891; CaM = EF651988; RPB2 = EF651941).
Colony diam, 7 d (mm): CYA 5–20; MEA 5–20; CY20S 10–26; CY20S 30 °C 0–20; CY20S 37 °C No growth; M40Y 48–70; M60Y 48–>75; M60Y 30 °C 44–>75; M60Y 37 °C No growth; CYAS 11–55; DG18 25–44; MEA10S 18–50.
Colony characters: CY20S 25 °C, 7 d: Colonies low to moderately deep, plane; margins entire; mycelium white or sulphur yellow (15) or orange (7); texture floccose; sporulation absent to sparse; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse luteous (12). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse luteous (12) or orange (7). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and sulphur yellow (15); texture floccose; sporulation sparse to moderately dense; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation absent to moderately dense; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse primrose (66) to luteous (12). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and sulphur yellow (15) or orange (7); texture floccose; sporulation sparse to moderately dense; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse primrose (66) or luteous (12). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium white or sulphur yellow (15) or orange (7); margins entire; texture floccose; sporulation absent to moderately dense; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse primrose (66) or luteous (12).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 100–240 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies smooth or slightly verruculose or rough along equatorial ridges, 4–6 × 3–5 μm, in side view lenticular, furrow present or pronounced, crests absent. Conidiophores with smooth stipes, hyaline or light brown, 250–1000 × 8–16.5 μm. Vesicles globose to subglobose, 20–50 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 6–12 × 3–5.5 μm. Conidia globose, subglobose to ellipsoidal, tuberculate, 5–7.5(–10) × 4–6(–7) μm. In culture ex-type (CBS 121.45) ascomata are absent, irregular proliferating conidiophores and phialides are produced, conidia measuring 9–17.5 × 7–13 μm.
Distinguishing characters: Phylogenetically A. proliferans is closely related to A. glaucus and A. aurantiacoflavus, but A. glaucus produces larger ascospores (5.5–7.5 × 3.5–6 μm) with irregular crests, A. aurantiacoflavus produces verruculose ascospores and has orange and yellow colony. The non-crested ascospores of A. proliferans resemble those of A. pseudoglaucus and A. ruber, but the latter two species grow well on M60Y at 37 °C, while A. proliferans does not grow under the same conditions.
Notes: Aspergillus proliferans was listed as the only anamorphic species in Aspergillus sect. Aspergillus by Thom & Raper (1945) and they observed the presence of cleistothecial initials (Fig. 40) which indicated some deficiency in the sexual cycle. The sexual strains were connected to this species by molecular data (Hubka et al., 2013a, Asgari et al., 2014), and are also confirmed by this study.
Additional materials examined: China, Tibet, Yak dung, CGMCC 3.04666. China, unknown source, CGMCC 3.04667, CGMCC 3.03971. China, Tibet, donkey dung, CGMCC 3.04668. China, Tibet, soil, CGMCC 3.04671. China, Hebei, soil, CGMCC 3.06523. China, Xinjiang, soil, CGMCC 3.10130. China, Yunnan, soil, CGMCC 3.06095. China, Yunnan, moldy wood, CGMCC 3.06495. China, Hebei, corn, CGMCC 3.04670. China, Hebei, unknown source, CGMCC 3.06097. Czech Republic, Prague, palm skin, 28-year-old woman, 2008, isolated by M. Skořepová, CCF 4096 = NRRL 62482 = DTO 355-C8. Czech Republic, Prague, toenail of 64-year-old man, 2010, isolated by P. Lysková, CCF 4115 = NRRL 62497 = DTO 355-C9. Czech Republic, Prague, toenail of 48-year-old man, 2011, isolated by P. Lysková, CCF 4146 = NRRL 62494 = DTO 355-D1. Czech Republic, Opava, stuffed bird, 2010, isolated by M. Polásek, CCF 4232. Czech Republic, Prague, toenail of 66-year-old man, 2011, isolated by P. Lysková, CCF 4263. South Korea, soybeans, 2012, isolated by D.H. Kim, CCF 4789 = KACC 47146 = DTO 355-D3. Spain, Andalusia, Aracena, Gruta de la Maravillas, cave sediment, 2010, isolated by A. Nováková , CCF 4192 = DTO 355-C6. The Netherlands, egg waffles, 2014, isolated by M. Meijer, DTO 322-A2. USA, Massachusetts, unknown source, NRRL 114 = DTO 355-C7 = CCF 5579. USA, Louisiana, library, inside the book, 2012, isolated by Ž. Jurjević, EMSL No. 1814. USA, Maryland, leafhoppers, isolated by V.K. Charles, NRRL 71 = DTO 355-D2 = CCF 5578. USA, Pennsylvania, Yardley, air of living room, 2013, isolated by Ž. Jurjević, EMSL No. 2207 = CCF 5395 = DTO 355-H5. USA, New York, Troy, basement, settle plates, 2015, isolated by Ž. Jurjević, EMSL No. 2791 = CCF 5392 = DTO 355-H6.
Aspergillus pseudoglaucus Blochwitz, Ann. Mycol. 27: 207. 1929. MycoBank MB275429. Fig. 42.
Fig. 42.
Aspergillus pseudoglaucus CBS 101747. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Synonyms: Eurotium pseudoglaucum Malloch & Cain, Canad. J. Bot. 50: 64. 1972.
Aspergillus glaucoaffinis Samson & W. Gams, Advances in Penicillium and Aspergillus Systematics: 47. 1985.
Eurotium repens var. pseudoglaucum (Blochwitz) Kozak., Mycol. Pap. 161: 76. 1989.
Eurotium repens de Bary, Hedwigia: 52. 1870.
Aspergillus reptans Samson & W. Gams, Advances in Penicillium and Aspergillus Systematics: 48. 1985.
Aspergillus glaber Blaser, Sydowia 28: 35. 1975.
Eurotium glabrum Blaser, Sydowia 28: 35. 1975.
Aspergillus fimicola H.Z. Kong & Z.T. Qi, Acta Mycol. Sin. 14 (2): 86. 1995.
Eurotium fimicola H.Z. Kong & Z.T. Qi, Acta Mycol. Sin. 14 (2): 86. 1995.
Typus: IMI 016122ii, lectotype (Samson & Gams 1985). Culture ex-type: CBS 123.28 = NRRL 40 = ATCC 10066 = IBT 5353 = IMI 016122 = IMI 016122ii = LSHBA 19 = MUCL 15624 = QM 7463 = Tom 5343 = WB 40.
ITS barcode: EF652050. (Alternative markers: BenA = EF651917; CaM = EF652007; RPB2 = EF651952).
Colony diam, 7 d (mm): CYA 20–35; MEA 19–26; CY20S 38–60; CY20S 30 °C 36–53; CY20S 37 °C No growth; M40Y 65–>75; M60Y 35–>75; M60Y 30 °C 53–>75; M60Y 37 °C 35–>75; CYAS 60–72; DG18 52–>75; MEA10S 50–65.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) or orange (7) and white; texture floccose; sporulation sparse to moderately dense; conidia en masse pale green (19) to yellow-green (71); soluble pigments absent; exudates absent; reverse straw (46), greenish olivaceous (90) or luteous (12). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) or orange (7); texture floccose; sporulation sparse to moderately dense; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse luteous (12) or fulvous (43). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse to moderately dense; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white or sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse greenish olivaceous (90); soluble pigments absent; exudates absent; reverse grey olivaceous (107) at centre, fading into light grey olivaceous (107). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and sulphur yellow (15) or orange (7); texture floccose; sporulation sparse or moderately dense; conidia en masse pale green (19) to dark greenish olivaceous (90); soluble pigments absent; exudates absent; reverse sulphur yellow (15) or ochreous (44). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium white or sulphur yellow (15) or orange (7); margins entire; texture floccose; sporulation sparse or moderately dense; conidia en masse pale green (19) to dark greenish olivaceous (90); soluble pigments absent; exudates absent; reverse ochreous (44).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 75–200 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies smooth or slightly rough, 4–6.5 × 3–4.5 μm, in side view lenticular, furrow absent or showing as a trace, crests absent. Conidiophores with smooth stipes, hyaline or light brown, 500–1 000 × (7–)11–22 μm. Vesicles globose to subglobose, (26–)37–65 μm wide (degenerated smaller vesicles measuring 11–21 μm were observed in ex-type CBS 123.28), fertile over two thirds to entire surface. Phialides flask-shaped, 6–11 × 4–6.5 μm. Conidia globose to subglobose, in most strains tuberculate; microtuberculate in atypical strain CBS 379.75, (3.5)–6–9 × (3–)5.5–7.5 μm.
Distinguishing characters: Morphologically A. pseudoglaucus resembles A. proliferans and A. ruber in ascospore size and ornamentation, however the ascospores of A. pseudoglaucus do not have or have indefinite furrow, while A. proliferans and A. ruber have more pronounced furrow on ascospores. The growth profile characters on M60Y at 37 °C can be used to distinguish A. pseudoglaucus and A. proliferans, the latter species cannot grow under this condition (Table 5). Aspergillus pseudoglaucus belongs to the A. ruber clade (Fig. 1). Other species in the A. ruber clade such as A. cumulatus, A. appendiculatus and A. mallochii can be differentiated by presence of crests, while A. tonophilus and A. sloanii do not grow on MEA and CYA at 25 °C and A. zutongqii has larger ascospores measuring 6–7.5 × 4.5–6 μm.
Notes: Aspergillus repens (de Bary) Fischer is a later homonym of Aspergillus repens (Corda) Sacc. 1882 pertaining to a different species, and A. pseudoglaucus is considered the correct name for Eurotium repens (Hubka et al. 2013a), we concur with this.
Additional materials examined: Canada, Quebec, cake, collected by A. Lafond, CBS 117314 = CCFC 008006 = DAOM 221134. China, Tibet, animal dung, CBS 101747 = CGMCC 3.04674 (AS 3.4674). China, tea, 1952, CGMCC 3.00460 = DTO 348-B9. China, Xinjiang, nest, CGMCC 3.06123. China, unknown source, CGMCC 3.01292, CGMCC 3.00452, CGMCC 3.00107, CGMCC 3.00472, CGMCC 3.03976, CGMCC 3.00456, CGMCC 3.01231, CGMCC 3.01070, CGMCC 3.03959, CGMCC 3.04063, CGMCC 3.00455, CGMCC 3.00461, CGMCC 3.00666, CGMCC 3.03565, CGMCC 3.03978, CGMCC 3.00133, CGMCC 3.01293. China, Sichuan, soil, CGMCC 3.06120. China, Hebei, unknown source, CGMCC 3.06112. China, Guangxi, earthworm faeces, CGMCC 3.06111. China, Yunnan, unknown source, CGMCC 3.06121. China, Zhejiang, soil, CGMCC 3.06110. China, Shandong, unknown source, CGMCC 3.06101. China, Yunnan, dry locust, CGMCC 3.06488. China, Yunnan, moldy noodles, CGMCC 3.06508. China, Shanxi, soil, CGMCC 3.06107. China, Heilongjiang, soil, CGMCC 3.06094. China, Fujiang, leaf, CGMCC 3.06105. China, Ningxia, soil, CGMCC 3.06079. China, Tibet, soil, CGMCC 3.06119. China, Neimenggu, unknown source, CGMCC 3.06117. China, Hebei, moldy mushroom, CGMCC 3.06505. China, Hebei, dung, CGMCC 3.06500. China, Yunnan, moldy herbs, CGMCC 3.06509. China, Guangdong, soil, CGMCC 3.06093. China, Hainan, coccid, CGMCC 3.06106. China, Hebei, soil, CGMCC 3.06502. China, Hebei, straw, CGMCC 3.06504. China, Beijing, unknown source, CGMCC 3.06115, CGMCC 3.06113, CGMCC 3.06114. China, Beijing, herbs, CGMCC 3.06080. Czech Republic, Prague, 2002, isolated by A. Kubátová, CCF 3283. Czech Republic, Prague, back skin of 39-year-old woman, 2008, isolated by M. Skořepová, CCF 4011. Czech Republic, Říčany, trunk skin of 39-year-old woman, 2009, isolated by P. Lysková and Z. Kolací, CCF 4072. Czech Republic, Prague, toenail of 57-year-old woman, 2011, isolated by P. Lysková and L. Jelínková, CCF 4372. Czech Republic, Prague, fingernail of 37-year-old man, 2011, isolated by P. Lysková and H.A. Macková, CCF 4373. Czech Republic, Prague, toenail of 31-year-old woman, 2007, isolated by M. Skořepová, CCF 4374. Czech Republic, near Mladeč Caves, outdoor air, 2012, isolated by A. Nováková, S86. France, Prunus domestica, isolated by da Fonseca, NRRL 13 = CBS 529.65. France, unknown source, isolated by A. Sartory, CBS 114.30. Hungary, indoor air, 2010, DTO 147-G3. Indonesia, Bali, tea, DTO 055-B3. Indonesia, Melastome, isolated by J. Houbraken, DTO 164-E5. Japan, Tokyo, unknown source, isolated by T. Ohtsuki, NRRL 25865. Nepal, Himalaya, soil, 1972, isolated by V. Janečková, CCF 1454. Portugal, unknown source, CBS 126221. Romania, Movile cave, Lake Room, Trachelipus troglobius faeces, 2011, isolated by A. Nováková, CCF 4950. Slovakia, Silická ladnica Cave, Archeological Dome, cave sediment, 2012, isolated by A. Nováková, S75. Spain, Madrid, chocolate, isolated by J. Varga, DTO 043-D3. Switzerland, Zuoz, Vaccinium myrtillus leaf, isolated by P. Blaser, CBS 379.75. Turkey, keratitis patient, isolated by M. Ilkit, DTO 244-D2. USA, wrist skin, NRRL 17. USA, Pennsylvania, floor swab, 2012, isolated by Ž. Jurjević, EMSL No. 1780 = CCF 5388 = DTO 355-I2. USA, Florida, Melbourne, vent, settle plates, 2015, isolated by Ž. Jurjević, EMSL No. 2779 = CCF 5389 = DTO 355-I3. USA, New York, Endicott, office, settle plates, 2015, isolated by Ž. Jurjević, EMSL No. 2809 = CCF 5386. USA, New Jersey, Piscataway, air, basement, 2014, isolated by Ž. Jurjević, EMSL No. 2474 = CCF 5387 = DTO 355-I4. USA, Missouri, St. Louis, cheddar cheese, 2015, isolated by Ž. Jurjević, EMSL No. 2853 = CCF 5390 = DTO 355-I5. The Netherlands, parmezan cheese, isolated by J. Houbraken, CBS 108961 = DTO 351-D2. USA, NY, Elmsford, swab, wallet drawer, 2014, isolated by Ž. Jurjević, EMSL No. 2643. USA, IL, Chicago, rubber toy import from China, 2015, isolated by Ž. Jurjević, EMSL No. 2695. USA, NY, Orangeburg, plastic bottle, 2015, isolated by Ž. Jurjević, EMSL No. 2789. USA, NY, Hempstead, living room, rug, 2013, isolated by Ž. Jurjević, EMSL No. 2190. USA, KY, Bowling Green, living room, air, 2015, isolated by Ž. Jurjević, EMSL No. 2862. Unknown country, milk powder, DTO 278-D5; quail egg, DTO 315-E8, DTO 315-E7; dolphin bones, 2010, isolated by T. Hoogenhuijzen, DTO 128-E8; gingerbread, DTO 235-B3.
Aspergillus ruber (Jos. König et al.) Thom & Church, Aspergillus: 112. 1926. MycoBank MB490579. Fig. 43.
Fig. 43.
Aspergillus ruber CBS 530.65T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Synonyms: Eurotium rubrum J. König, Spieck. & W. Bremer, Z. Untersuch. Nahr. u. Genussm. 4: 726. 1901.
Aspergillus rubrobrunneus Samson & W. Gams, Advances in Penicillium and Aspergillus Systematics: 49. 1985.
Aspergillus athecius Raper & Fennell, The Genus Aspergillus: 183. 1965.
Gymnoeurotium athecium (Raper & Fennell) Malloch & Cain, Canad. J. Bot. 50 (12): 2619. 1972.
Edyuillia athecia (Raper & Fennell) Subram., Curr. Sci. 41: 756. 1972.
Eurotium athecium (Raper & Fennell) Arx, The genera of fungi sporulating in pure culture: 91. 1974.
Aspergillus atheciellus Samson & W. Gams, Advances in Penicillium and Aspergillus Systematics: 34. 1985.
Aspergillus tuberculatus Z.T. Qi & Z.M. Sun, Acta Mycol. Sin. 13: 86. 1994.
Eurotium tuberculatum Z.T. Qi & Z.M., Acta Mycol. Sin. 13: 86. 1994.
Typus: CBS 530.65, neotype (Samson & Gams 1985). Culture ex-type: CBS 530.65 = NRRL 52 = ATCC 16441 = IBT 5453 = IMI 211380 = JCM 22942 = QM 1973 = Thom 5599B = WB 52.
ITS barcode: EF652066. (Alternative markers: BenA = EF651920; CaM = EF652009; RPB2 = EF651947).
Colony diam, 7 d (mm): CYA 21–22; MEA 15–16; CY20S 51–52; CY20S 30 °C 18–30; CY20S 37 °C No growth; M40Y >75; M60Y >75; M60Y 30 °C >75; M60Y 37 °C >75; CYAS 65–66; DG18 >75; MEA10S 65–67.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse green (20) or vinaceous buff (86); soluble pigments absent; exudates absent; reverse sulphur yellow (15) at centre, pale green (20) at edge. M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) later turning orange (7); texture floccose; sporulation moderately dense; conidia en masse green (20) or vinaceous buff (86); soluble pigments absent; exudates absent; reverse ochreous (44). M60Y 25 °C, 7 d: Colonies deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse greyish green (50) or vinaceous buff (86); soluble pigments absent; exudates absent; reverse ochreous (44). CYAS 25 °C, 7 d: Colonies deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation dense; conidia en masse greyish green (50) or vinaceous buff (86); soluble pigments absent; exudates absent; reverse buff (45). DG18 25 °C, 7 d: Colonies deep, plane; margins entire; mycelium white and sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse greyish green (50) or vinaceous buff (86); soluble pigments absent; exudates absent; reverse amber (47). MEA10S 25 °C, 7 d: Colonies deep, plane; mycelium white and sulphur yellow (15); margins entire; texture floccose; sporulation moderately dense; conidia en masse greyish green (50) or vinaceous buff (86); soluble pigments absent; exudates absent; reverse ochreous (44).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 50–175 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies in most strains smooth or minute rough along equatorial ridges, tuberculate in atypical strain CBS 101748, 4–6 × 3.5–5 μm, in side view lenticular, furrow present or pronounced, crests absent. Conidiophores with smooth stipes, hyaline or light brown, 500–750 × 7–13.5 μm. Vesicles globose to subglobose, 25–48 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 7–9(–12) × 3.5–6 μm. Conidia subglobose to ellipsoidal, tuberculate, (4.5–)7–9(–12) × 4–6(–8) μm.
Distinguishing characters: Phylogenetically, Aspergillus ruber is closely related to A. zutongqii, but A. zutongqii produces larger ascospores measuring 6–7.5 × 4.5–6 μm. Morphologically, A. ruber resembles A. proliferans in ascospore and conidia morphology, but A. proliferans cannot grow on M60Y at 37 °C.
Additional materials examined: Argentina, Buenos Aires Prov., San Martin, honey sample, 2007, isolated by M.C. Hostench, CBS 123575. Brazil, Corn kernels, 2008, isolated by J. Houbraken, DTO 060-I9. Canada, British Columbia, hay, collected by V. Chang, CBS 117310. China, Shanxi, soil, CBS 101748 = CGMCC 3.04632 (AS 3.4632). China, tea, 1952, CGMCC 3.00457 = DTO 348-B6. China, tea, CGMCC 3.00458. China, unknown source CGMCC3.02577, CGMCC 3.02573, CGMCC 3.00459, CGMCC 3.03957, CGMCC 3.01296, CGMCC 3.00401, CGMCC 3.01298, CGMCC 3.01297, CGMCC 3.01069, CGMCC 3.00439, CGMCC 3.00388, CGMCC 3.04318, CGMCC 3.04315, CGMCC 3.04061, CGMCC 3.00298, CGMCC 3. 01295. China, Beijing, medicinal herb, CGMCC 3.06125. China, Beijing, beverage, CGMCC 3.09054. China, Beijing, unknown source, CGMCC 3.06130, CGMCC 3.06127, CGMCC 3.06129. China, Shanxi, Wugong, soil, CGMCC 3.06137. China, Henan, unknown source, CGMCC 3.06124. China, pig hair, CGMCC 3.03551. China, Hebei, soil, CGMCC 3.06497. China, Hainan, resin, CGMCC 3.06118. China, Xinjiang, nest, CGMCC 3.06122. China, Hunan, unknown source, CGMCC 3.06098. China, Hebei, straw, CGMCC 3.06499. China, Shanxi, Wugong, soil, CGMCC 3.04632. China, Pu'er tea, isolated by J. Houbraken, DTO 257-G7. Czech Republic, Nymburk, malt dust, 1993, isolated by A. Kubátová, CCF 2920. Czech Republic, Prague, toenail of 60-year-old woman, 2011, isolated by P. Lysková, CCF 4377. Czech Republic, Prague, Coptish textile (Museum of Decorative Arts), 1999, A. Kubátová, CCF 3464. Czech Republic, white pepper, isolated by L. Marvanová, CCM F-438. Czech Republic, Prague, toenail of 32-year-old man, 2010, isolated by P. Lysková, CCF 4104. Germany, archive, 2009, isolated by J. Houbraken, DTO 088-E3. Indonesia, peanuts, 2008, isolated by J. Houbraken, DTO 062-I5, DTO 062-I9, DTO 063-A2. Indonesia, Geography Library (stacks), 2012, isolated by Rahmawati, from air in Yogyakarta, DTO 238-C4. Thailand, coffee beans, 2006, isolated by P. Noonim, DTO 287-A1, DTO 287-A2, DTO 289-A6, DTO 286-E5. UK, coffee beans, 1965, isolated by E. Yuill, NRRL 5000 = CBS 464.65. Zaire, leaf, isolated by J. Houbraken, DTO 257-F8. Unknown source, isolated by G. Pollacci, CBS 110.31. Unknown country, tobacco, isolated by M. Meijer, DTO 220-A9. Unknown source, isolated by G. Smith, NRRL 76. Unknown source, 1918, isolated by O. Goethals, CBS 104.18 = DTO 351-C4.
Aspergillus sloanii Visagie, Hirooka & Samson, Stud. Mycol. 78: 108. 2014. MycoBank MB809194. Fig. 44.
Fig. 44.
Aspergillus sloanii CBS 138177T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Typus: CBS H-21811, holotype. Culture ex-type: CBS 138177 = DTO 245-A1 = IBT 34509.
ITS barcode: KJ775540. (Alternative markers: BenA = KJ775074; CaM = KJ775309; RPB2 = KX463365).
Colony diam, 7 d (mm): CYA 2–4; MEA No growth; CY20S 9–15; CY20S 30 °C No growth; CY20S 37 °C No growth; M40Y 46–67; M60Y 55–>75; M60Y 30 °C 47–61; M60Y 37 °C No growth; CYAS 17–27; DG18 28–55; MEA10S 40–55.
Colony characters: CY20S 25 °C, 7 d: Colonies low, plane; margins entire; mycelium white and straw (46); texture floccose; sporulation sparse; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse white. M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) and white; texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) and white; texture floccose; sporulation sparse; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) and white; texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) and white; texture floccose; sporulation sparse; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse sulphur yellow (15). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium sulphur yellow (15) and white; margins entire; texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 60–205 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies smooth, minute rough along equatorial ridges, 4–6 × 3–4.5 μm, in side view lenticular, furrow present, crests absent. Conidiophores with smooth stipes, hyaline or light brown, 160–900 × 7.5–16 μm. Vesicles globose to subglobose, (10–)34–53 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, (7.5–)9–13.5(–18) × (5–)7–9.5 μm. Conidia globose, tuberculate, 5.5–9.5 × 5.5–9 μm.
Distinguishing characters: Aspergillus sloanii resembles A. ruber, A. proliferans and A. pseudoglaucus in ascospore morphology, A. sloanii does not grow or grows very restrictedly on CYA and MEA. Good growth occurs on M40Y and M60Y.
Additional materials examined: UK, Middlesex, house dust, 2010, isolated by E. Whitfield & K. Mwange, CBS 138176 = DTO 244-I8 = CCF 4926, CBS 138231 = DTO 245-A6, CBS 138178 = DTO 245-A8, CBS 138179 = DTO 245-A9.
Aspergillus tamarindosoli A.J. Chen, Frisvad & Samson, sp. nov. MycoBank MB818737. Fig. 45.
Fig. 45.
Aspergillus tamarindosoli CBS 141775T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Etymology: Name refers to its origin, isolated from soil under tamarind.
Diagnosis: Verruculose ascospores with 0.5–1 μm crests, wide vesicles measuring 40–72 μm, lobate-reticulate conidia measuring 4–7 × 3–4.5 μm.
Typus: Thailand, Hua Hin, soil under tamarind, 2007, isolated by R. Samson & J. Houbraken (holotype CBS H-22826, culture ex-type: CBS 141775 = DTO 054-A8 = IBT 34432).
ITS barcode: LT670981. (Alternative markers: BenA = LT671191; CaM = LT671192; RPB2 = LT671193).
Colony diam, 7 d (mm): CYA 16–17; MEA 13–15; CY20S 40–43; CY20S 30 °C 14–16; CY20S 37 °C No growth; M40Y >75; M60Y >75; M60Y 30 °C >75; M60Y 37 °C 40–45; CYAS 40–42; DG18 35–36; MEA10S 46–48.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse pale green (19) to greyish green (50); soluble pigments absent; exudates absent; reverse sulphur yellow (15). M40Y 25 °C, 7 d: Colonies moderately deep, slightly sulcate; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse pale green (19) to greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse pale green (19) to greyish green (50); soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse pale green (19) to greyish green (50); soluble pigments absent; exudates absent; reverse primrose (66). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44) at centre, fading into yellow-green (71). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium white; margins entire; texture floccose; sporulation dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 130–240 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies verruculose, 3.5–5 × 3–4 μm, in side view lenticular, furrow present, crests irregular, 0.5–1.5 μm. Conidiophores with smooth stipes, hyaline or light brown, 700–1 000 × 10–15 μm. Vesicles globose to subglobose, 40–72 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 6.5–12 × 4–5.5 μm. Conidia subglobose to ellipsoidal, lobate-reticulate, 4–7 × 3–4.5 μm.
Distinguishing characters: Aspergillus tamarindosoli resembles A. chevalieri in ascospore morphology, but A. chevalieri produces smaller conidia measuring 3–4(–6) × 2.5–3.5(–5) μm and narrower vesicles measuring 23–47 μm.
Aspergillus teporis A.J. Chen, Frisvad & Samson, sp. nov. MycoBank MB818738. Fig. 46.
Fig. 46.
Aspergillus teporis CBS 141768T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Etymology: Name refers to its origin, isolated from heat treated corn kernels.
Diagnosis: Protuberance presented on ascospore convex and furrow.
Typus: The Netherlands, heat treated corn kernels, 2008, isolated by M. Meijer (holotype CBS H-22821, culture ex-type: CBS 141768 = DTO 058-E5 = IBT 34513).
ITS barcode: LT670982. (Alternative markers: BenA = LT671194; CaM = LT671195; RPB2 = LT671196).
Colony diam, 7 d (mm): CYA 19–20; MEA 16–18; CY20S 46–47; CY20S 30 °C 48–50; CY20S 37 °C 49–50; M40Y 53–56; M60Y 50–54; M60Y 30 °C 55–63; M60Y 37 °C >75; CYAS 28–29; DG18 30–37; MEA10S 35–40.
Colony characters: CY20S 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46); texture floccose; sporulation sparse; conidia en masse pale green (19) to greyish green (50); soluble pigments absent; exudates absent; reverse straw (46). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46); texture floccose; sporulation sparse; conidia en masse greyish green (50) to dark green (21); soluble pigments absent; exudates absent; reverse luteous (12). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) and white; texture floccose; sporulation sparse; conidia en masse greyish green (50) to dark green (21); soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) and white; texture floccose; sporulation sparse; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse straw (46). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46); texture floccose; sporulation sparse; conidia en masse greyish green (50) to dark green (21); soluble pigments absent; exudates absent; reverse straw (46). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium straw (46); margins entire; texture floccose; sporulation sparse; conidia en masse greyish green (50) to dark green (21); soluble pigments absent; exudates absent; reverse luteous (12).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, cream yellow, globose to subglobose, 120–180 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies slightly verruculose, 5–6.5 × 4–5.5 μm, in side view lenticular, furrow pronounced, with scattered protuberance, crests 0.5 μm. Conidiophores with smooth stipes, hyaline or light brown, 800–1 200 × 8–19 μm. Vesicles globose to subglobose, 33–53 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 7–12 × 3.5–5 μm. Conidia globose, subglobose to ellipsoidal, lobate-reticulate, 3.5–6 × 3–4.5 μm.
Distinguishing characters: Aspergillus teporis is a representative of the basal clades of sect. Aspergillus. It is closely related to A. leucocarpus, which produces white ascomata and larger conidia (5.5–9 × 5–8 μm). Under the SEM, the protuberance present on the ascospore convex and furrow can distinguish A. teporis from other taxa in this section.
Aspergillus tonophilus Ohtsuki, Bot. Mag. (Tokyo) 75: 438. 1962. MycoBank MB326663. Fig. 47.
Fig. 47.
Aspergillus tonophilus KACC 47150. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Synonyms: Eurotium tonophilum Ohtsuki, Bot. Mag. (Tokyo) 75: 438. 1962.
Typus: IMI 108299, neotype (Samson & Gams 1985). Culture ex-type: CBS 405.65 = NRRL 5124 = ATCC 16440 = ATCC 36504 = IBT 21230 = IMI 108299 = QM 8599 = WB 5124.
ITS barcode: EF652081. (Alternative markers: BenA = EF651919; CaM = EF652000; RPB2 = EF651969).
Colony diam, 7 d (mm): CYA No growth; MEA No growth; CY20S 24–25; CY20S 30 °C No growth; CY20S 37 °C No growth; M40Y >75; M60Y >75; M60Y 30 °C >75; M60Y 37 °C 8–9; CYAS 49–53; DG18 56–58; MEA10S 56–58.
Colony characters: CY20S 25 °C, 7 d: Colonies low, plane; margins entire; mycelium sulphur yellow (15) and white; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse white. M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse luteous (12). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse luteous (12). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse luteous (12) fading to sulphur yellow (15). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation moderately dense; conidia en masse pale green (19); soluble pigments absent; exudates absent; reverse sulphur yellow (15). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium sulphur yellow (15); margins entire; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse luteous (12).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 100–235 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies verruculose, 4–6 × 3–4.5 μm, in side view lenticular, furrow present, crests absent. Conidiophores with smooth stipes, hyaline or light brown, 120–500 × 7–12.5 μm. Vesicles globose to subglobose, 25–44 μm wide (degenerated, smaller vesicles measuring 8–16 μm were observed in ex-type CBS 405.65), fertile over two thirds to entire surface. Phialides flask-shaped, 6–11 × 3–5 μm. Conidia globose to subglobose, tuberculate to lobate-reticulate, 5–7.5 × 3.5–6 μm.
Distinguishing characters: Aspergillus tonophilus is a member of A. ruber clade (Fig. 1). The colonies of A. tonophilus remain brightly yellow even after two weeks of cultivation in contrast to other species from the A. ruber clade. The ascospores of A. tonophilus resemble those of A. aurantiacoflavus, however, A. aurantiacoflavus produces orange and yellow colonies and slightly larger conidia measuring 5–9 × 4–7 μm.
Additional materials examined: Canada, house dust, 2015, isolated by C.M. Visagie, DTO 356-H6 = KAS6175. South Korea, meju, 2012, isolated by S.B. Hong, KACC 45365 = CCF 4785 = DTO 355-A2. South Korea, soybeans, 2012, isolated by D.H. Kim, KACC 47150 = CCF 4786 = DTO 355-A1.
Aspergillus xerophilus Samson & Mouch., Antonie van Leeuwenhoek 41: 348. 1975. MycoBank MB309251. Fig. 48.
Fig. 48.
Aspergillus xerophilus CBS 938.73T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Synonyms: Eurotium xerophilum Samson & Mouch, Antonie van Leeuwenhoek 41: 348. 1975.
Typus: CBS 938.73, holotype. Culture ex-type: CBS 938.73 = NRRL 6131 = IBT 5429 = IBT 5489 = IBT 34503 = DTO 083-A2.
ITS barcode: EF652085. (Alternative markers: BenA = EF651923; CaM = EF651983; RPB2 = EF651970).
Colony diam, 7 d (mm): CYA No growth; MEA No growth; CY20S No growth; CY20S 30 °C No growth; CY20S 37 °C No growth; M40Y 60–62; M60Y >75; M60Y 30 °C 65–>75; M60Y 37 °C No growth; CYAS No growth; DG18 39–55; MEA10S 67–>75.
Colony characters: M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) and olivaceous buff (89) at centre; texture floccose; sporulation sparse; soluble pigments absent; exudates absent; reverse luteous (12). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) and olivaceous buff (89) at centre; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse luteous (12). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium straw (46) and white; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse luteous (12). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium straw (46) and white; margins entire; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse ochreous (44).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 165–330 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies verruculose, 4.5–6.5 × 3.5–5 μm, in side view lenticular, furrow present, crests irregular, < 0.5 μm. Conidiophores with smooth stipes, hyaline or light brown, 50–200 × 6.5–9.5–(12) μm. Vesicles globose to subglobose, 40–66 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 6–9 × 3.5–6 μm. Conidia globose to subglobose, microtuberculate, 3.5–5.5 × 3–4.5 μm.
Distinguishing characters: Phylogenetically A. xerophilus is closely related to A. osmophilus, but A. osmophilus produces larger ascospores (7–9 × 6–7.5 μm) and grows on M60Y at 37 °C. Morphologically A. xerophilus resembles A. endophyticus in ascospore ornamentation, but the ascospores of A. endophyticus have longer crests (0.5–1 μm).
Additional materials examined: Egypt, Western desert, desert soil, isolated by J. Mouchacca, NRRL 6132 = CBS 755.74.
Aspergillus zutongqii A.J. Chen, Frisvad & Samson, sp. nov. MycoBank MB818739. Fig. 49.
Fig. 49.
Aspergillus zutongqii CBS 141773T. A. Colonies after 7 d at 25 °C: top row left to right, CYA, M40Y, CY20S and CYAS; bottom row left to right, MEA, reverse M40Y, M60Y and DG18. B, E. Conidiophores. C, D. Ascomata. F, I. Ascospores. G, H. Conidia. Scale bars: B, C = 20 μm; D = 250 μm; E–G = 10 μm; H, I = 2 μm.
Etymology: Named in honour of Chinese mycologist Zutong Qi, who wrote first Aspergillus monograph in China, and contributed his whole career to Aspergillus taxonomy in China.
Diagnosis: Large, verruculose, non-crested ascospores measuring 6–7.5 × 4.5–6 μm.
Typus: China, Beijing, peanut shell, 2008, isolated by L. Wang (holotype CBS H-22824, culture ex-type: CBS 141773 = CGMCC 3.13917 = DTO 349-E1 = IBT 34450).
ITS barcode: LT670986. (Alternative markers: BenA = LT671206; CaM = LT671207; RPB2 = LT671208).
Colony diam, 7 d (mm): CYA 14–15; MEA 7–17; CY20S 33–38; CY20S 30 °C 13–20; CY20S 37 °C No growth; M40Y >75; M60Y >75; M60Y 30 °C >75; M60Y 37 °C 10–30; CYAS 32–50; DG18 42–50; MEA10S 56–60.
Colony characters: CY20S 25 °C, 7 d: Colonies low to moderately deep, plane; margins entire; mycelium sulphur yellow (15) or ochreous (44); texture floccose; sporulation absent or sparse, conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44). M40Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) and orange (7); texture floccose; sporulation sparse to moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44) to orange (7). M60Y 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) and orange (7); texture floccose; sporulation sparse to moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44) to orange (7). CYAS 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) and orange (7); texture floccose; sporulation sparse to moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44) to umber (9). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) and orange (7); texture floccose; sporulation sparse to moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44) to orange (7). MEA10S 25 °C, 7 d: Colonies moderately deep, plane; mycelium sulphur yellow (15); margins entire; texture floccose; sporulation sparse to moderately dense; conidia en masse greyish green (50); soluble pigments absent; exudates absent; reverse ochreous (44) to orange (7).
Micromorphology: Ascomata eurotium-like, cleistothecial, superficial, yellow, globose to subglobose, 110–220 μm. Asci 8 spored, globose to subglobose. Ascospores hyaline, in surface view globose to subglobose, spore bodies verruculose, 6–7.5 × 4.5–6 μm, in side view lenticular, furrow pronounced, crests absent. Conidiophores with smooth stipes, hyaline or light brown, 150–500 × 7.5–13 μm. Vesicles globose to subglobose, 25–40 μm wide, fertile over two thirds to entire surface. Phialides flask-shaped, 8–12 × 4–6.5 μm. Conidia subglobose to ellipsoidal, tuberculate, 5.5–10 × 4–7 μm.
Distinguishing characters: Phylogenetically and morphologically, Aspergillus zutongqii is close to A. ruber, but A. ruber produces smaller ascospores (4–6 × 3.5–5 μm).
Additional materials examined: China, Ningxia, 2001, CGMCC 3.06103 = DTO 348-F7. China, 1969, isolated by Z.T. Qi, CGMCC 3.03980 = DTO 348-D7. China, ocular lens, 1969, isolated by Z.T. Qi, CGMCC 3.03961 = DTO 348-D5.
Notes
Aspergillus taklimakanensis Abliz & Y. Horie, Mycoscience 42: 289. 2001. MycoBank MB474683
Synonyms: Eurotium taklimakanense Abliz & Y. Horie, Mycoscience 42: 289. 2001.
This species was accepted by Guarro et al. (2012), but was treated as invalid by Hubka et al. (2013a), because the holotype CBM-FA-876 includes different species (probably A. cristatus), and is in conflict with the protologue. No living culture or herbarium material corresponding with the protologue is extant.
Acknowledgements
This project was supported by the Alfred P. Sloan Foundation Program on the Microbiology of the Built Environment (grant number G-2014-14529), by the Hungarian Research Fund (OTKA K115690), by the project of the Charles University Grant Agency (GAUK 1434217) and the project BIOCEV (CZ.1.05/1.1.00/02.0109) provided by the Ministry of Education, Youth and Sports of CR and ERDF, by the National Natural Science Foundation of China No. 81473345. We thank Miroslav Kolařík for his support and advice, Milada Chudíčková for her invaluable assistance in the laboratory, CCF collection staff (Ivana Kelnarová, Adéla Kovaříčková) for deposition and lyophilization of the cultures, Miroslav Hyliš for assistance with scanning electron microscopy. We thank Bingda Sun, Lei Wang, Seung-Beom Hong, Ivana Kelnarová, Ondřej Koukol, Alena Nováková, Pavlína Lysková, Magdalena Skořepová, Vanda Chrenková, Naďa Mallátová and Polona Zalar for providing some interesting cultures. Vit Hubka is grateful for support from the Czechoslovak Microscopy Society (CSMS scholarship 2016).
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Contributor Information
A.J. Chen, Email: amanda_j_chen@163.com.
R.A. Samson, Email: r.samson@westerdijkinstitute.nl.
References
- Abe K., Nagao Y., Nakada T. Assessment of indoor climate in an apartment by use of a fungal index. Applied and Environmental Microbiology. 1996;62:959–963. doi: 10.1128/aem.62.3.959-963.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A.M., Ismail S.A., Abd-El-Rahman H.A. Quantitative, qualitative and toxigenic evaluations of xerophilic mold in traditional Egyptian salted fish, Molouha. Journal of Food Safety. 2005;25:9–18. [Google Scholar]
- Al-Julaifi M.Z. Ochratoxin A production by Eurotium amstelodami and Eurotium spp. isolated from locally grown barley in Saudi Arabia. Kuwait Journal of Science and Engineering. 2003;30:59–66. [Google Scholar]
- Allen C.M., Jr. Isoprene-containing metabolites of Aspergillus amstelodami. Canadian Journal of Microbiology. 1972;18:1275–1282. doi: 10.1139/m72-197. [DOI] [PubMed] [Google Scholar]
- Almeida A.P., Dethoup T., Singburaudom N. The in vitro anticancer activity of the crude extract of the sponge-associated fungus Eurotium cristatum and its secondary metabolites. Journal of Natural Pharmaceuticals. 2010;1:25–29. [Google Scholar]
- Amend A.S., Seifert K.A., Samson R. Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. PNAS. 2010;107:13748–13753. doi: 10.1073/pnas.1000454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anke H., Kolthoum I., Laatsch H. Metabolic products of microorganisms. 192. The anthraquinones of the Aspergillus glaucus group. II. Biological activity. Archives of Microbiology. 1980;126:231–236. doi: 10.1007/BF00409925. [DOI] [PubMed] [Google Scholar]
- Anke H., Kolthoum I., Zähner H. Metabolic products of microorganisms. 185. The anthraquinones of the Aspergillus glaucus group. I. Occurrence, isolation, identification and antimicrobial activity. Archives of Microbiology. 1980;126:223–230. doi: 10.1007/BF00409924. [DOI] [PubMed] [Google Scholar]
- Anke H., Zähner H., Koenig W. Metabolic products of microorganisms. 170. On the antibiotic activity of cladosporin. Archives of Microbiology. 1978;116:253–257. doi: 10.1007/BF00417848. [DOI] [PubMed] [Google Scholar]
- Anslow W.K., Raistrick H. Studies in the biochemistry of microorganisms. 67. The molecular constitutions of catenarin and erythroglaucin, metabolic products respectively of Helminthosporium catenarium Drechsler and of species in the Aspergillus glaucus series. Biochemical Journal. 1940;34:1124–1133. doi: 10.1042/bj0341124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai K., Aoki Y., Yamamoto Y. Asperinines A and B, dimeric tetrahydroanthracene derivatives from Aspergillus ruber. Chemical and Pharmaceutical Bulletin. 1989;37:621–625. [Google Scholar]
- Asgari B., Zare R., Zamanizadeh H.R. Aspergillus osmophilus sp. nov., and a new teleomorph for A. proliferans. Mycoscience. 2014;55:53–62. [Google Scholar]
- Ashley J.N., Raistrick H., Richards T. Studies in the biochemistry of microorganisms. LXII. The crystalline colouring matters of species in the Aspergillus glaucus series. Biochemical Journal. 1939;33:1291–1303. doi: 10.1042/bj0331291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assante G., Camarda L., Nasini G. Secondary mould metabolites. IX. Structure of a new bianthrone and three new secoanthraquinones from Aspergillus wentii Wehmer. Gazzetta Chimica Italiana. 1980;110:629–631. [Google Scholar]
- Bachmann M., Blaser P., Lüthy J. Toxicity and mutagenicity of anthraquinones from Aspergillus chevalieri. Journal of Environmental Pathology, Toxicology and Oncology. 1982;11:113–116. [PubMed] [Google Scholar]
- Bachmann M., Lüthy J., Schlatter C. Toxicity and mutagenicity of molds of the Aspergillus glaucus group. Identification of physcion and three related anthraquinones as main toxic constituents from Aspergillus chevalieri. Journal of Agricultural and Food Chemistry. 1979;27:1342–1347. doi: 10.1021/jf60226a021. [DOI] [PubMed] [Google Scholar]
- Barbetta M., Casnati G., Pochini A. Neoechinuline: a new indole metabolite from Aspergillus amstelodami. Tetrahedron Letters. 1969;10:4457–4460. [Google Scholar]
- Baudisch C., Assadian O., Kramer A. Concentration of the genera Aspergillus, Eurotium and Penicillium in 63-μm house dust fraction as a method to predict hidden moisture damage in homes. BMC Public Health. 2009;9:247. doi: 10.1186/1471-2458-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J.W., Klich M. Mycotoxins. Clinical Microbiology Reviews. 2003;16:497–516. doi: 10.1128/CMR.16.3.497-516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benny G.L., Kimbrough J.W. A synopsis of the orders and families of Plectomycetes with keys to genera. Mycotaxon. 1980;12:1–91. [Google Scholar]
- Birch A.J. The origin of the C5-unit in auroglaucin. Chemistry and Industry. 1958;1958:1321. [Google Scholar]
- Blaser P. Taxonomische und physiologische Untersuchungen über die Gattung Eurotium Link ex Fries. Sydowia. 1975;28:1–49. [Google Scholar]
- Blaser P., Ramstein H., Schmidt-Lorenz W. Toxicität und Mutagenität der xerophilen Schimmelpilze der Gattung Eurotium (Aspergillus glaucus gruppe) Lebensmittel-Wissenschaft & Technologie. 1980;14:66–71. [Google Scholar]
- Büchi G., Klaubert D.H., Shank R.C. Structure and synthesis of kotanin and desmethylkotanin, metabolites of Aspergillus glaucus. Journal of Organic Chemistry. 1971;36:1143–1147. doi: 10.1021/jo00807a028. [DOI] [PubMed] [Google Scholar]
- Burkin A.A., Kononenko G.P. Producers of mycophenolic acid in ensiled and grain feeds. Applied Biochemistry and Microbiology. 2010;46:592–598. [PubMed] [Google Scholar]
- Cardillo R., Fuganti C., Gatti G. Molecular structure of cryptoechinuline A, a new metabolite of Aspergillus amstelodami, isolated during investigation of echinuline biosynthesis. Tetrahedron Letters. 1974;15:3163–3166. [Google Scholar]
- Cardillo R., Fuganti C., Ghiringhelli D. Stereochemical course of the α,β-desaturation of L-tryptophan in the biosynthesis of cryptoechinuline A in Aspergillus amstelodami. Journal of the Chemical Society, Chemical Communications. 1975:778–779. [Google Scholar]
- Cattel L., Grove J.F., Shaw D. New metabolic products of Aspergillus flavus. Part III. Biosynthesis of asperentin. Journal of the Chemical Society, Perkin Transactions 1. 1973:2626–2629. doi: 10.1039/p19730002626. [DOI] [PubMed] [Google Scholar]
- Chelkowski J., Samson R.A., Wiewiórowska M. Ochratoxin A formation by isolated strains of the conidial state of Aspergillus glaucus Link ex Grey (= Eurotium herbariorum Wiggers Link ex Gray) from cereal grains. Nahrung. 1987;31:267–269. doi: 10.1002/food.19870310402. [DOI] [PubMed] [Google Scholar]
- Chen G.D., Bao Y.R., Huang Y.F. Three pairs of variecolortide enantiomers from Eurotium sp. with caspase-3 inhibitory activity. Fitoterapia. 2014;92:252–259. doi: 10.1016/j.fitote.2013.11.012. [DOI] [PubMed] [Google Scholar]
- Chen A.J., Frisvad J.C., Sun B.D. Aspergillus section Nidulantes (formerly Emericella): polyphasic taxonomy, chemistry and biology. Studies in Mycology. 2016;84:1–118. doi: 10.1016/j.simyco.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen C.M., Papavizas G.C., Benjamin C.R. A new halophilic species of Eurotium. Mycologia. 1959;51:636–640. [Google Scholar]
- Cochrane R.V.K., Sanichar R., Lambkin G.R. Production of new cladosporin analogues by reconstitution of the polyketide synthases responsible for the biosynthesis of this antimalarial agent. Angewandte Chemie International Edition. 2016;55:664–668. doi: 10.1002/anie.201509345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coveney R.D., Peck H.M., Townsend R.J. Recent advances in mycotoxicosis. Society of Chemical Industry (London) 1966;23:31–43. [Google Scholar]
- Cox R.E., Chexal K.K., Holker J.S.E. The biosynthesis of fungal metabolites. Part VIII. Identification of N-benzoyl-L-phenylalanyl-L-phenylalaniol acetate, a metabolite of Aspergillus glaucus. Journal of the Chemical Society, Perkin Transactions 1. 1976:578–580. doi: 10.1039/p19760000578. [DOI] [PubMed] [Google Scholar]
- de Hoog G.S., Gerrits van den Ende A.H. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses. 1998;41:183–189. doi: 10.1111/j.1439-0507.1998.tb00321.x. [DOI] [PubMed] [Google Scholar]
- de Hoog G.S., Guarro G., Gené J. 2nd edn. Centraalbureau voor Schimmelcultures (CBS); Utrecht, the Netherlands: 2000. Atlas of clinical fungi. [Google Scholar]
- Dimici L., Wada S. Lipid changes in bonito meat in the katsuobushi processing and quality assessment of the commercial product based on lipid composition. Journal of Japan Oil Chemists' Society. 1994;43:470–478. [Google Scholar]
- Dossena A., Marchelli R., Pochini A. New metabolites of Aspergillus amstelodami related to the biogenesis of neoechinulin. Journal of the Chemical Society, Chemical Communications. 1974:771–772. [PubMed] [Google Scholar]
- Du F.Y., Li X.M., Li C.S. Cristatumin A–D, new indole alkaloids from the marine-derived endophytic fungus Eurotium cristatum EN-220. Bioorganic and Medicinal Chemistry Letters. 2012;22:4650–4653. doi: 10.1016/j.bmcl.2012.05.088. [DOI] [PubMed] [Google Scholar]
- Du F.Y., Li X.M., Song J.Y. Anthraquinone derivatives and an orsellinic acid ester from the marine alga-derived endophytic fungus Eurotium cristatum EN-220. Helvetica Chimica Acta. 2014;97:973–978. [Google Scholar]
- Du L., Ai J., Li D. Aspergiolides C and D: spirocyclic aromatic polyketides with potent protein kinase c-Met inhibitory effects. Chemistry – A European Journal. 2011;17:1319–1326. doi: 10.1002/chem.201001547. [DOI] [PubMed] [Google Scholar]
- Du L., Zhu T., Fang Y. Aspergiolide A, a novel anthraquinone derivative with naphtho[1,2,3-de]chromene-2,7-dione skeleton isolated from a marine-derived fungus Aspergillus glaucus. Tetrahedron. 2007;63:1085–1088. [Google Scholar]
- Du L., Zhu T.J., Liu H.B. Cytotoxic polyketides from a marine-derived fungus Aspergillus glaucus. Journal of Natural Products. 2008;71:1837–1842. doi: 10.1021/np800303t. [DOI] [PubMed] [Google Scholar]
- El-Kady I., El-Maraghy S., Zohri A.N. Mycotoxin producing potential of some isolates of Aspergillus flavus and Eurotium groups from meat products. Microbiological Research. 1994;149:297–307. doi: 10.1016/S0944-5013(11)80073-X. [DOI] [PubMed] [Google Scholar]
- Ellestad G.A., Kunstmann M.P., Mirando P. Structures of fungal diterpene antibiotics LL-S491β and -γ. Journal of the American Chemical Society. 1972;94:6206–6208. doi: 10.1021/ja00772a054. [DOI] [PubMed] [Google Scholar]
- Ellestad G.A., Mirando P., Kuntsmann M.P. Structure of the metabolite LL-S490β from an unidentified Aspergillus species. Journal of Organic Chemistry. 1973;38:4204–4205. doi: 10.1021/jo00963a024. [DOI] [PubMed] [Google Scholar]
- Engstrom G.W., Stenkamp R.E., McDorman D.J. Spectral identification, X-ray structure determination, and iron-chelating capability of erythroglaucin, a red pigment from Aspergillus ruber. Journal of Agricultural and Food Chemistry. 1982;30:304–307. [Google Scholar]
- Fraga M.E., Curvello F., Gatti M.J. Potential aflatoxin and ochratoxin A production by Aspergillus species in poultry feed processing. Veterinary Research Communications. 2007;31:343–353. doi: 10.1007/s11259-006-3434-x. [DOI] [PubMed] [Google Scholar]
- Fraga M.E., Direito G.M., Gatti M.J. Revaluation of aflatoxin production by Aspergillus candidus and Eurotium chevalieri isolated from poultry feed in Brazil. Revista Brasileira de Medicina Veterinaria. 2008;30:86–90. [Google Scholar]
- Frisvad J.C., Larsen T.O. Extrolites of Aspergillus fumigatus and other pathogenic species in Aspergillus section Fumigati. Frontiers in Microbiology. 2016;6:1485. doi: 10.3389/fmicb.2015.01485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad J.C., Thrane U. Standardized high-performance liquid chromatography of 182 mycotoxins and other fungal metabolites based on alkylphenone indices and UV-VIS spectra (diode-array detection) Journal of Chromatography. 1987;404:195–214. doi: 10.1016/s0021-9673(01)86850-3. [DOI] [PubMed] [Google Scholar]
- Frisvad J.C., Thrane U. Liquid column chromatography of mycotoxins. In: Betina V., editor. Chromatography of mycotoxins: techniques and applications. Elsevier; Amsterdam: 1993. pp. 253–372. (Journal of Chromatography Library 54). [Google Scholar]
- Frisvad J.C., Thrane U., Samson R.A. Mycotoxin producers. In: Dijksterhuis J., Samson R.A., editors. Food mycology. A multifaceted approach to fungi and food. CRC Press; Boca Raton, Florida: 2007. pp. 135–159. [Google Scholar]
- Fujimoto H., Fujimaki T., Okuyama E. Immunomodulatory constituents from an ascomycete, Microascus tardifaciens. Chemical and Pharmaceutical Bulletin. 1999;47:1426–1432. doi: 10.1248/cpb.47.1426. [DOI] [PubMed] [Google Scholar]
- Gams W., Christensen M., Onions A.H.S. Infrageneric taxa of Aspergillus. In: Samson R.A., Pitt J.I., editors. Advances in Penicillium and Aspergillus systematics. Vol. 102. Plenum Press; New York: 1985. pp. 55–62. (NATO ASI Series. Ser. A.: Life Sciences). [Google Scholar]
- Gao H., Liu W., Zhu T. Diketopiperazine alkaloids from a mangrove rhizosphere soil derived fungus Aspergillus effuses H1-1. Organic and Biomolecular Chemistry. 2012;10:9501–9506. doi: 10.1039/c2ob26757h. [DOI] [PubMed] [Google Scholar]
- Gao H., Zhu T., Li D. Prenylated indole diketopiparazine alkaloids from a mangrove rhizosphere soil derived fungus Aspergillus effuses H1-1. Archives of Pharmacal Research. 2013;36:952–956. doi: 10.1007/s12272-013-0107-5. [DOI] [PubMed] [Google Scholar]
- Gao J., León F., Radwan M.M. Benzyl derivatives with in vitro binding affinity for human opioid and cannabinoid receptors from the fungus Eurotium repens. Journal of Natural Products. 2011;74:1636–1639. doi: 10.1021/np200147c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Radwan M.M., León F. Antimicrobial and antiprotozoal activities of secondary metabolites from the fungus Eurotium repens. Medicinal Chemistry Research. 2012;21:3080–3086. doi: 10.1007/s00044-011-9798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti G., Cardillo R., Fuganti C. Molecular structure of cryptoechinuline G, and isoprenylated dehydrotryptophan metabolite isolated from Aspergillus ruber. Tetrahedron Letters. 1978;19:2605–2606. [Google Scholar]
- Gatti G., Cardillo R., Fuganti C. Structure determination of two extractives from Aspergillus amstelodami by nuclear magnetic resonance spectroscopy. Journal of the Chemical Society, Chemical Communications. 1976:435–436. [Google Scholar]
- Gatti G., Fuganti C. NMR spectra of echinulin and related compounds. Journal of Chemical Research. 1979;11:366–367. [Google Scholar]
- Glass N.L., Donaldson G.C. Development of premier sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Applied and Environmental Microbiology. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes N.M., Dethoup T., Singburaudom N. Eurocristatine, a new diketopiperazizne dimer from the marine sponge-associated fungus Eurotium cristatum. Phytochemistry Letters. 2012;5:717–720. [Google Scholar]
- González-Andrade M., Del Valle P., Macías-Rubalcava M.L. Calmodulin inhibitors from Aspergillus stromatioides. Chemistry & Biodiversity. 2013;10:328–336. doi: 10.1002/cbdv.201200321. [DOI] [PubMed] [Google Scholar]
- Gould B.S., Raistrick H. Studies in the biochemistry of microorganisms. XL. The crystalline pigments of species in the Aspergillus glaucus series. Biochemical Journal. 1934;28:1640–1656. doi: 10.1042/bj0281640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco M., Kemppainen M., Pose G. Taxonomic characterization and secondary metabolite profiling of Aspergillus section Aspergillus contaminating feeds and feedstuffs. Toxins. 2015;7:3512–3537. doi: 10.3390/toxins7093512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J.F. New metabolic products of Aspergillus flavus. Part II. Asperflavin, anhydroasperflavin, and 5,7-dihydroxy-4-methylphthalide. Journal of the Chemical Society, Perkin Transactions 1. 1972:2406–2411. doi: 10.1039/p19720002406. [DOI] [PubMed] [Google Scholar]
- Grove J.F. New metabolic products of Aspergillus flavus. I. Asperentin, its methyl ethers, and 5′-hydroxyasperentin. Journal of the Chemical Society, Perkin Transactions 1. 1972:2400–2406. doi: 10.1039/p19720002400. [DOI] [PubMed] [Google Scholar]
- Grove J.F. New metabolic products of Aspergillus flavus. IV. 4′-hydroxyasperentin and 5′-hydroxyasperentin-8-methyl ether. Journal of the Chemical Society, Perkin Transactions 1. 1973:2704–2706. doi: 10.1039/p19730002704. [DOI] [PubMed] [Google Scholar]
- Guarro J., Gené J., Stchigel A.M. CBS-KNAW Fungal Biodiversity Centre; Utrecht, the Netherlands: 2012. Atlas of soil ascomycetes. CBS Biodiversity Series 10. [Google Scholar]
- Hamasaki T., Fukunaga M., Kimura Y. Isolation and structures of two new metabolites from Aspergillus ruber. Agricultural and Biological Chemistry. 1980;44:1685–1687. [Google Scholar]
- Hamasaki T., Kimura Y., Hatsuda Y. Structure of a new metabolite, dihydroauroglaucin, produced by Aspergillus chevalieri. Agricultural and Biological Chemistry. 1981;45:313–314. [Google Scholar]
- Hamasaki T., Nagayama K., Hatsuda Y. A new metabolite, L-alanyl-L-tryptophan anhydride from Aspergillus chevalieri. Agricultural and Biological Chemistry. 1976;40:2487. [Google Scholar]
- Hamasaki T., Nagayama K., Hatsuda Y. Structure of a new metabolite from Aspergillus chevalieri. Agricultural and Biological Chemistry. 1976;40:203–205. [Google Scholar]
- Hayakawa K., Ueno Y., Nakanishi S. Production of fish sauce from fish meal treated with koji-mould. Seibutsu Kogaku Kaishi. 1993;71:245–251. [Google Scholar]
- Hong S.B., Go S.J., Shin H.D. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia. 2005;97:1316–1329. doi: 10.3852/mycologia.97.6.1316. [DOI] [PubMed] [Google Scholar]
- Hong S.B., Kim D.H., Lee M. Taxonomy of Eurotium species isolated from Meju. The Journal of Microbiology. 2011;49:669–674. doi: 10.1007/s12275-011-0376-y. [DOI] [PubMed] [Google Scholar]
- Hong S.B., Lee M., Kim D.H. Aspergillus cibarius sp. nov. from traditional meju in Korea. The Journal of Microbiology. 2012;50:712–714. doi: 10.1007/s12275-012-2347-3. [DOI] [PubMed] [Google Scholar]
- Horie Y., Abliz P., Hui Y. Emericella qinqixianii, a new species from desert soil in China. Mycoscience. 2000;41:183–187. [Google Scholar]
- Houbraken J., de Vries R.P., Samson R.A. Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Advances in Applied Microbiology. 2014;86:199–249. doi: 10.1016/B978-0-12-800262-9.00004-4. [DOI] [PubMed] [Google Scholar]
- Houbraken J., Samson R.A. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology. 2011;70:1–51. doi: 10.3114/sim.2011.70.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V., Kolařík β-tubulin paralogue tubC is frequently misidentified as the benA gene in Aspergillus section Nigri taxonomy: primer specificity testing and taxonomic consequences. Persoonia. 2012;29:1–10. doi: 10.3767/003158512X658123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V., Kolařík M., Kubátová A. Taxonomical revision of Eurotium and transfer of species to Aspergillus. Mycologia. 2013;105:912–937. doi: 10.3852/12-151. [DOI] [PubMed] [Google Scholar]
- Hubka V., Kubatova A., Mallatova N. Rare and new etiological agents revealed among 178 clinical Aspergillus strains obtained from Czech patients and characterised by molecular sequencing. Medical Mycology. 2012;50:601–610. doi: 10.3109/13693786.2012.667578. [DOI] [PubMed] [Google Scholar]
- Hubka V., Nováková A., Kolařík M. Revision of Aspergillus section Flavipedes: seven new species and proposal of section Jani sect. nov. Mycologia. 2015;107:169–208. doi: 10.3852/14-059. [DOI] [PubMed] [Google Scholar]
- Hubka V., Nováková A., Samson R.A. Aspergillus europaeus sp. nov., a widely distributed soil-borne species related to A. wentii (section Cremei) Plant Systematics and Evolution. 2016;302:641–650. [Google Scholar]
- Hubka V., Peterson S.W., Frisvad J.C. Aspergillus waksmanii sp. nov. and Aspergillus marvanovae sp. nov., two closely related species in section Fumigati. International Journal of Systematic Evolutionary Microbiology. 2013;63:783–789. doi: 10.1099/ijs.0.047076-0. [DOI] [PubMed] [Google Scholar]
- Inoue S., Hashizume K., Takamatsu N. Synthetic studies on echinulin and related products. IV. Isolation, structure and synthesis of flavoglaucin-auroglaucin type natural products isolated from Aspergillus amstelodami. Yakugaku Zasshi. 1977;97:569–575. doi: 10.1248/yakushi1947.97.5_569. [DOI] [PubMed] [Google Scholar]
- Inoue S., Murata J., Takamatsu N. Synthetic studies on echinulin and related products. V. Isolation, structure and synthesis of echinulin-neoechinulin type alkaloids isolated from Aspergillus amstelodami. Yakugaku Zasshi. 1977;97:576–581. doi: 10.1248/yakushi1947.97.5_576. [DOI] [PubMed] [Google Scholar]
- Inoue S., Takamatsu N., Hashizume K. Synthetic studies on echinulin and related natural products. VI. Structure and synthesis of aurechinulin. Yakugaku Zasshi. 1977;97:582–585. doi: 10.1248/yakushi1947.97.5_582. [DOI] [PubMed] [Google Scholar]
- Ishikawa Y., Morimoto K., Hamasaki T. Flavoglaucin, a metabolite of Eurotium chevalieri, its antioxidation and synergism with tocopherol. Journal of the American Oil Chemists' Society. 1984;61:1864–1868. [Google Scholar]
- Ishikawa Y., Morimoto K., Hamasaki T. Metabolites of Eurotium species, their antioxidative properties and synergism with tocopherol. Journal of Food Science. 1985;50:1742–1744. [Google Scholar]
- Itabashi T., Matsuishi N., Hosoe T. Two new dioxopiperazine derivatives, arestricticin A and B, isolated from Aspergillus restrictus and Aspergillus penicillioides. Chemical and Pharmaceutical Bulletin. 2006;54:1639–1641. doi: 10.1248/cpb.54.1639. [DOI] [PubMed] [Google Scholar]
- Jayraman P., Kalyanasundaram I. Natural occurrence of toxigenic fungi and mycotoxin in rice bran. Mycopathologia. 1990;110:81–85. doi: 10.1007/BF00446995. [DOI] [PubMed] [Google Scholar]
- Jurjević Ž., Kubátová A., Kolařík M. Taxonomy of Aspergillus section Petersonii sect. nov. encompassing indoor and soil-borne species with predominant tropical distribution. Plant Systematics and Evolution. 2015;301:2441–2462. [Google Scholar]
- Karo M., Hadlok R. Investigations on sterigmatocystin production by fungi of the genus Eurotium. In: Krogh P., editor. International IUPAC Symposium on Mycotoxins and Phycotoxins. Technical University of Vienna; Vienna, Austria: 1982. pp. 178–181. [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Kim S.H., Kwon S.W. Aspergillus cumulatus sp. nov. from rice straw and air for meju fermentation. Journal of Microbiology and Biotechnology. 2014;24:334–336. doi: 10.4014/jmb.1312.12006. [DOI] [PubMed] [Google Scholar]
- Kimura Y., Shimomura N., Tanigawa F. Plant growth activities of aspyran, asperentin, and its analogues produced by the fungus Aspergillus sp. Zeitschrift für Naturforschung C. 2012;67:587–593. doi: 10.1515/znc-2012-11-1209. [DOI] [PubMed] [Google Scholar]
- Klich M.A. Centraalbureau voor Schimmelcultures; Utrecht, the Netherlands: 2002. Identification of common Aspergillus species. [Google Scholar]
- Kocsubé S., Perrone G., Magistà D. Aspergillus is monophyletic: evidence from multiple gene phylogenies and extrolites profiles. Studies in Mycology. 2016;85:199–213. doi: 10.1016/j.simyco.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong H.Z., Qi Z.T. Two new species of Eurotium Link. Acta Mycologica Sinica. 1995;14:10–16. [Google Scholar]
- Kozakiewicz Z. Aspergillus species on stored products. Mycological Papers. 1989;161:1–188. [Google Scholar]
- Kozlovsky A.G., Zhelifonova V.P., Antipova T.Y. Exo-metabolites of mycelial fungi isolated in production premises of cheese-making and meat-processing plants. Food Additives and Contaminants Part A. 2014;31:300–306. doi: 10.1080/19440049.2013.862350. [DOI] [PubMed] [Google Scholar]
- Kulik M.M., Holaday C.E. Aflatoxin: a metabolic product of several fungi. Mycopathologia et Mycologia Applicata. 1966;30:137–140. doi: 10.1007/BF02130360. [DOI] [PubMed] [Google Scholar]
- Kuntze O. Vol. 2. Arthur Felix; Germany, Leipzig: 1891. pp. 375–1011. (Revisio Generum Plantarum). [Google Scholar]
- Kuttruff C.A., Zipse H., Trauner D. Concise total synthesis of variecolortide A and B through an unusual hetero-Diels-Alder reaction. Angewandte Chemie International Edition. 2011;50:1402–1405. doi: 10.1002/anie.201006154. [DOI] [PubMed] [Google Scholar]
- Laatsch H., Anke H. Stoffwechselprodukte von Microorganismen, 214. Viocristin, isoviocristin und hydroxyviocristin. struktur und synthese natürlich vorkommenden 1,4-anthraquinone. Liebigs Annalen der Chemie. 1982:2189–2215. [Google Scholar]
- Lanfear R., Calcott B., Ho S.Y.W. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- Leitao J., LeBars J., Bailly J.R. Production of aflatoxin B1 production by Aspergillus ruber Thom and Church. Mycopathologia. 1989;108:135–138. doi: 10.1007/BF00436064. [DOI] [PubMed] [Google Scholar]
- Li D.L., Li X.M., Li T.G. Benzaldehyde derivatives from Eurotium rubrum, an endophytic fungus derived from the mangrove plant Hibiscus tiliaceus. Chemical and Pharmaceutical Bulletin. 2008;56:1282–1285. doi: 10.1248/cpb.56.1282. [DOI] [PubMed] [Google Scholar]
- Li D.L., Li X.M., Li T.G. Dioxopiperazine alkaloids produced by the marine mangrove derived endophytic fungus Eurotium rubrum. Helvetica Chimica Acta. 2008;91:1888–1892. [Google Scholar]
- Li D.L., Li X.M., Proksch P. 7-O-methylvariecolortide A, a new spirocyclic diketopiperazine alkaloid from a marine mangrove derived endophytic fungus, Eurotium rubrum. Natural Product Communications. 2010;5:1583–1586. [PubMed] [Google Scholar]
- Li D.L., Li X.M., Wang B.G. Natural anthraquinone derivatives from a marine mangrove plant-derived endophytic fungus Eurotium rubrum: structural elucidation and PPPH radical scavenging activity. Journal of Microbiology and Biotechnology. 2009;19:675–680. [PubMed] [Google Scholar]
- Li Y., Li X., Kang J.S. New radical scavenging and ultraviolet-A protecting prenylated dioxopiperazine alkaloid related to isoechinulin A from a marine isolate of the fungus Aspergillus. The Journal of Antibiotics. 2004;57:337–340. doi: 10.7164/antibiotics.57.337. [DOI] [PubMed] [Google Scholar]
- Li Y., Li X., Kim S.K. Golmaneone, a new diketopiperazine alkaloid from the marine-derived fungus Aspergillus sp. Chemical and Pharmaceutical Bulletin. 2004;52:375–376. doi: 10.1248/cpb.52.375. [DOI] [PubMed] [Google Scholar]
- Li Y., Li X., Lee U. A new radical scavenging anthracene glycoside, asperflavin ribofuranoside, and polyketides from a marine isolate of fungus Microsporum. Chemical and Pharmaceutical Bulletin. 2006;54:882–883. doi: 10.1248/cpb.54.882. [DOI] [PubMed] [Google Scholar]
- Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Malloch D., Cain R.F. The Trichocomataceae: ascomycetes with Aspergillus, Paecilomyces and Penicillium imperfect states. Canadian Journal of Botany. 1972;50:2613–2628. [Google Scholar]
- Mangin M.L. What is Aspergillus glaucus? Critical and experimental study of the form grouped under this name. Annales des Sciences Naturelles. Botanique. 1909;10:303–371. [Google Scholar]
- Marchelli R., Dossena A., Casnati G. Biosynthesis of neoechinulin by Aspergillus amstelodami from cyclo-L-[U-14C]alanyl-L-[5,7-3H2]tryptophyl. Journal of the Chemical Society, Chemical Communications. 1975:779–780. [Google Scholar]
- Marchelli R., Dossena A., Pochini A. The structures of five new didehydropeptides related to neoechinulin, isolated from Aspergillus amstelodami. Journal of the Chemical Society, Perkin Transactions 1. 1977:713–717. [PubMed] [Google Scholar]
- Masclaux F., Guého E., de Hoog G.S. Phylogenetic relationships of human-pathogenic Cladosporium (Xylohypha) species inferred from partial LS rRNA sequences. Medical Mycology. 1995;33:327–338. doi: 10.1080/02681219580000651. [DOI] [PubMed] [Google Scholar]
- McNeill J., Barrie F.R., Buck W.R. Koeltz Scientific Books; Königstein: 2012. International code of nomenclature for algae, fungi, and plants (Melbourne Code) adopted by the Eighteenth International Botanical Congress, Melbourne, Australia, July 2011. Regnum Vegetabile 154. [Google Scholar]
- Meng L.H., Du F.Y., Li X.M. Rubrumazine A–C, indolediketopiperazines of the isoechinulin class from Eurotium rubrum MA-150, a fungus obtained from marine mangrove-derived rhizospheric soil. Journal of Natural Products. 2015;78:909–913. doi: 10.1021/np5007839. [DOI] [PubMed] [Google Scholar]
- Meng L.H., Mándi A., Li X.M. Isolation, stereochemical study, and antioxidant activity of benzofuranone derivatives from a mangrove-derived fungus Eurotium rubrum MA-150. Chirality. 2016;28:581–584. doi: 10.1002/chir.22613. [DOI] [PubMed] [Google Scholar]
- Micheluz A., Sulyok M., Manente S. Fungal secondary metabolite analysis applied to cultural heritage: the case of a contaminated library in Venice. World Mycotoxin Journal. 2016;9:397–407. [Google Scholar]
- Miyake Y., Ito C., Itoigawa M. Antioxidants produced by Eurotium herbariorum of filamentous fungi used for the manufacture of karebushi, dried bonito (katsuobushi) Bioscience Biotechnology and Biochemistry. 2009;73:1323–1327. doi: 10.1271/bbb.80887. [DOI] [PubMed] [Google Scholar]
- Miyake Y., Ito C., Kimura T. Isolation of aromatic compounds produced by Eurotium herbariorum NU-2 from Karebushi, a katsuobushi, and their DPPH-radical scavenging activities. Food Science and Technology Research. 2014;20:139–146. [Google Scholar]
- Miyake Y., Ito C., Tokuda H. Evaluation of flavoglaucin, its derivatives and pyranonigrin produced by molds used in fermented foods for inhibiting tumor promotion. Bioscience Biotechnology and Biochemistry. 2010;74:1120–1122. doi: 10.1271/bbb.90955. [DOI] [PubMed] [Google Scholar]
- Miyake Y., Mochizuki M., Ito C. Peroxynitrite scavengers produced by filamentous fungus used in the katsuobushi manufacturing process. Food Science and Technology Research. 2010;16:493–498. [Google Scholar]
- Moubasher A.H., El-Kady I.A., Shoriet A. Toxigenic aspergilli isolated from different sources in Egypt. Annales de la Nutrition et de l'Alimentation. 1977;31:607–615. [PubMed] [Google Scholar]
- Nagasawa H., Isdogai A., Suzuki A. Structures of isoechinulins A, B and C, new indole metabolites from Aspergillus ruber. Tetrahedron Letters. 1976;17:1601–1604. [Google Scholar]
- Nagasawa H., Usogai A., Ikeda K. Isolation and structure elucidation of a new indole metabolite from Aspergillus ruber. Agricultural and Biological Chemistry. 1975;39:1901–1902. [Google Scholar]
- Nakashima R., Slater G.P. The configuration of echinulin Part III. The absolute configuration of echinulin. Tetrahedron Letters. 1971;12:2649–2650. [Google Scholar]
- Nguyen L.T., Schmidt H.A., von Haeseler A. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K.F., Månsson M., Rank C. Dereplication of microbial natural products by LC-DAD-TOFMS. Journal of Natural Products. 2011;74:2338–2348. doi: 10.1021/np200254t. [DOI] [PubMed] [Google Scholar]
- Nielsen K.F., Mogensen J.M., Johansen M. Review of secondary metabolites and mycotoxins from the Aspergillus niger group. Analytical and Bioanalytical Chemistry. 2009;395:1225–1246. doi: 10.1007/s00216-009-3081-5. [DOI] [PubMed] [Google Scholar]
- O'Donnell K., Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- Oeemig J.S., Lynggaard C., Knudsen D.H. Eurocin, a new fungal defensin. The Journal of Biological Chemistry. 2012;287:42361–42372. doi: 10.1074/jbc.M112.382028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.W. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia. 2008;100:205–226. doi: 10.3852/mycologia.100.2.205. [DOI] [PubMed] [Google Scholar]
- Peterson S.W., Jurjević Ž. Talaromyces columbinus sp. nov., and genealogical concordance analysis in Talaromyces clade 2a. PLoS One. 2013;8(10):e78084. doi: 10.1371/journal.pone.0078084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.W., Varga J., Frisvad J.C. Phylogeny and subgeneric taxonomy of Aspergillus. In: Varga J., Samson R.A., editors. Aspergillus in the genomic era. Wageningen Academic Publishers; Wageningen: 2008. pp. 33–56. [Google Scholar]
- Pitt J.I. Nomenclatorial and taxonomic problems in the genus Eurotium. In: Samson R.A., Pitt J.I., editors. Advances in Penicillium and Aspergillus systematics. Vol. 102. Plenum Press; New York: 1985. pp. 383–396. (NATO ASI Series. Ser. A.: Life Sciences). [Google Scholar]
- Pitt J.I., Hocking A.D. Aspergillus and related teleomorphs. In: Pitt J., Hocking A.D., editors. Fungi and food spoilage. Springer; London: 2009. pp. 275–337. [Google Scholar]
- Pitt J.I., Samson R.A. Types of Aspergillus and Penicillium and their teleomorphs in current use. In: Samson R.A., Pitt J.I., editors. Integration of modern taxonomic methods for Penicillium and Aspergillus. Harwood Academic Publishers; Amsterdam, the Netherlands: 2000. pp. 51–72. [Google Scholar]
- Pitt J.I., Taylor J.W. Aspergillus, its sexual states, and the new International Code of Nomenclature. Mycologia. 2014;105:1051–1062. doi: 10.3852/14-060. [DOI] [PubMed] [Google Scholar]
- Pitt J.I., Taylor J.W. (2441) Proposal to conserve the name Aspergillus (Fungi: Eurotiales: Trichocomaceae) with a conserved type to maintain the name Eurotium. Taxon. 2016;65:631–632. [Google Scholar]
- Podojil M., Sedmera P., Vokoun J. Eurotium (Aspergillus) repens metabolites and their biological activity. Folia Microbiologica. 1979;23:438–443. doi: 10.1007/BF02885572. [DOI] [PubMed] [Google Scholar]
- Qi Z.T., Sun C.M. Identification of predominant species in brick tea. Acta Mycologica Sinica. 1990;9:176–179. [Google Scholar]
- Quilico A., Cardini C. The diffusion of echinulin in molds of the group Aspergillus glaucus. Atti della Accademia Nazionale dei Lincei, Classe di Scienze Fisiche, Matematiche e Naturali, Rendiconti Lincei Matematica E Applicazioni. 1950;9:220–228. [Google Scholar]
- Quilico A., Panazzi L. Chemische Untersuchungen über Aspergillus echinulatus. I. Mitteilung. Chemische Berichte. 1943;76:348–358. [Google Scholar]
- Quilico A., Panizzi L., Mugnaini E. Structure of flavoglaucin and auroglaucin. Nature. 1949;164:26–27. doi: 10.1038/164026a0. [DOI] [PubMed] [Google Scholar]
- Rabie C.J., Steyn P.S., van Heerden F.R. The isolation and identification of some toxic constituents of Aspergillus wentii Wehmer. Mycotoxin Research. 1986;2:19–24. doi: 10.1007/BF03191958. [DOI] [PubMed] [Google Scholar]
- Rank C., Nielsen K.F., Larsen T.O. Distribution of sterigmatocystin in filamentous fungi. Fungal Biology. 2011;115:406–420. doi: 10.1016/j.funbio.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Raper K.B., Fennell D.I. Williams & Wilkins; Baltimore, MD: 1965. The genus Aspergillus. [Google Scholar]
- Raper K.B. Nomenclature in Aspergillus and Penicillium. Mycologia. 1957;49:644–662. [Google Scholar]
- Rayner R.W. CMI and British Mycological Society; Kew, Surrey, England: 1970. A mycological colour chart. [Google Scholar]
- Réblová M., Hubka V., Thureborn O. From the tunnels into the treetops: new lineages of black yeasts from biofilm in the Stockholm metro system and their relatives among ant-associated fungi in the Chaetothyriales. PLoS One. 2016;11(10):e0163396. doi: 10.1371/journal.pone.0163396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboux G., Piarroux R., Mauny F. Role of molds in farmer's lung disease in eastern France. American Journal of Respiratory and Critical Care Medicine. 2001;163:1534–1539. doi: 10.1164/ajrccm.163.7.2006077. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel S., Reboux G., Dalphin K. Microbiological evolution of hay and relapse in patients with farmer's lung. Occupational and Environmental Medicine. 2004;61:e3. [PMC free article] [PubMed] [Google Scholar]
- Samson R.A. A compilation of the Aspergillus described since 1965. Studies in Mycology. 1979;18:1–38. [Google Scholar]
- Samson R.A., Gams W. Typification of the species of Aspergillus and associated teleomorphs. In: Samson R.A., Pitt J.I., editors. Advances in Penicillium and Aspergillus systematics. Vol. 102. Plenum Press; New York: 1985. pp. 31–54. (NATO ASI Series. Ser. A.: Life Sciences). [Google Scholar]
- Samson R.A., Houbraken J., Frisvad J.C. CBS-KNAW Fungal Biodiversity Centre; Utrecht: 2010. Food and indoor fungi. [Google Scholar]
- Samson R.A., Visagie C.M., Houbraken J. Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology. 2014;78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder H.W., Kelton W.H. Production of sterigmatocystin by some species of the genus Aspergillus and its toxicity to chicken embryos. Applied and Environmental Microbiology. 1975;30:589–591. doi: 10.1128/am.30.4.589-591.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séguin V., Gente S., Heutte N. First report of mycophenolic acid production by Eurotium repens isolated from agricultural and indoor environments. World Mycotoxin Journal. 2014;7:321–328. [Google Scholar]
- Semeniuk G., Nagel C.M., Gilman J.C. Observation on mold development and on deterioration in stored yellow dent shelled corn. Agricultural Experiment Station. Iowa State College of Agriculture. Research Bulletin. 1947;349:253–284. [Google Scholar]
- Shu Y.Z., Cutrone J.F.Q., Klohr S.E. BMS-192548, a tetracyclic binding inhibitor of neuropeptide Y, receptors from Aspergillus niger WB2346. 2. Physicochemical properties and structural characterization. The Journal of Antibiotics. 1995;48:1060–1065. doi: 10.7164/antibiotics.48.1060. [DOI] [PubMed] [Google Scholar]
- Šimonovičová A., Kraková L., Pangallo D. Fungi on mummified human remains and in the indoor air in the Kuffner family crypt in Sládkovičovo (Slovakia) International Biodeterioration & Biodegradation. 2015;99:157–164. [Google Scholar]
- Sklenář F., Jurjević Ž., Zalar P. Phylogeny of osmophillic aspergilli (subgenus Aspergillus) and taxonomic revision of section Restricti. Studies in Mycology. 2017 doi: 10.1016/j.simyco.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack G.J., Puniani E., Frisvad J.C. Secondary metabolites from Eurotium species, Aspergillus calidoustus and A. insuetus common in Canadian homes with a review of their chemistry and biological activities. Mycological Research. 2009;113:480–490. doi: 10.1016/j.mycres.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Smetanina O.F., Kalinovskii A.I., Khudyakova Y.V. Metabolites from the marine fungus Eurotium repens. Chemistry of Natural Compounds. 2007;43:395–398. [Google Scholar]
- Smith G. The effect of adding trace elements to Czapek-Dox medium. Transactions of the British Mycological Society. 1949;32:280–283. [Google Scholar]
- Soboleva N.A., Kurmanov I.A. Biosynthesis of sterigmatocystin. Veterinarya (Moscow) 1984;1:65–66. [Google Scholar]
- Stipanovic R.D., Schroeder H.W. Preechinulin, a metabolite of Aspergillus chevalieri. Transactions of the British Mycological Society. 1976;66:178–179. [Google Scholar]
- Stipanovics R.D., Schroeder H.W., Hein H. Identification of D-valyl-L-tryptophan anhydride from Aspergillus chevalieri. Lloydia Journal of Natural Products. 1976;39:158–159. [PubMed] [Google Scholar]
- Summerbell R.C., Cooper E., Bun U. Onychomycosis: a critical study of techniques and criteria for confirming the etiologic significance of nondermatophytes. Medical Mycology. 2005;43:39–59. doi: 10.1080/13693780410001712043. [DOI] [PubMed] [Google Scholar]
- Sun S.W., Ji C.Z., Gu Q.Q. Three new polyketides from marine-derived fungus Aspergillus glaucus HB1-19. Journal of Asian Natural Products Research. 2013;15:956–961. doi: 10.1080/10286020.2013.826205. [DOI] [PubMed] [Google Scholar]
- Sun X., Zhou X., Cai M. Identified biosynthetic pathway of aspergiolide A and a novel strategy to increase its production in a marine-derived fungus Aspergillus glaucus by feeding of biosynthetic precursors and inhibitors simultaneously. Bioresource Technology. 2009;100:4244–4251. doi: 10.1016/j.biortech.2009.03.061. [DOI] [PubMed] [Google Scholar]
- Sun Z.M., Qi Z.T. New taxa and a new record of Aspergillus and Eurotium. Acta Mycologica Sinica. 1994;13:81–87. [Google Scholar]
- Szebiotko K., Chelkowski J., Dopierala B. Mycotoxins in cereal grains Part I. Ochratoxin, citrinin, sterigmatocystin, penicillic acid and toxigenic fungi in cereal grain. Nahrung. 1981;25:415–421. [PubMed] [Google Scholar]
- Talice R.V., Mackinnon J.E. Aspergillus (Eurotium) montevidensis, n. sp. isolé d'un cas d'otomycose chez l'homme. Comptes rendus des séances de la Société de biologie et de ses filiales Société de biologie (France) 1931;108:1007–1009. [Google Scholar]
- Tang Q., Guo K., Li X.Y. Three new asperentin derivatives from the algicolous fungus Aspergillus sp. F00785. Marine Drugs. 2014;12:5993–6002. doi: 10.3390/md12125993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K., Du L., Sun X. Biosynthesis of aspergiolide A, a novel antitumor compound by a marine-derived fungus Aspergillus glaucus via the polyketide pathway. Tetrahedron Letters. 2009;50:1082–1085. [Google Scholar]
- Thom C., Raper K.B. The Aspergillus glaucus group. U.S. Department of Agriculture Miscellaneous Publications. 1941;426:1–46. [Google Scholar]
- Thom C., Raper K.B. Williams & Wilkins; Maryland, MD: 1945. A manual of the Aspergilli. [Google Scholar]
- Thrasher J.D. Fungi, bacteria, nano-particulates, mycotoxins and human health in water damaged indoor environments. Journal of Community and Public Health Nursing. 2016;2:115. [Google Scholar]
- Varga J., Due M., Frisvad J.C. Taxonomic revision of Aspergillus section Clavati based on molecular, morphological and physiological data. Studies in Mycology. 2007;59:89–106. doi: 10.3114/sim.2007.59.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J., Frisvad J.C., Samson R.A. A reappraisal of fungi producing aflatoxins. World Mycotoxin Journal. 2009;2:263–277. [Google Scholar]
- Visagie C.M., Hirooka Y., Tanney J.B. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Studies in Mycology. 2014;78:63–139. doi: 10.1016/j.simyco.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie C.M., Houbraken J., Frisvad J.C. Identification and nomenclature of the genus Penicillium. Studies in Mycology. 2014;78:343–371. doi: 10.1016/j.simyco.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie C.M., Houbraken J., Rodriques C. Five new Penicillium species in section Sclerotiora: a tribute to the Dutch Royal family. Persoonia. 2013;31:42–62. doi: 10.3767/003158513X667410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie C.M., Yilmaz N., Renaud J.B. A survey of xerophilic Aspergillus from indoor environment, including descriptions of two new section Aspergillus species producing eurotium-like sexual states. MycoKeys. 2017;19:1–30. [Google Scholar]
- Wang W.L., Liu P.P., Zhang Y.P. 2-hydroxydiplopterol, a new cytotoxic pentacyclic triterpenoid from the halotolerant fungus Aspergillus variecolor B-17. Archives of Pharmacal Research. 2009;32:1211–1214. doi: 10.1007/s12272-009-1904-8. [DOI] [PubMed] [Google Scholar]
- Wang W.L., Lu Z.Y., Tao H.W. Isoechinulin-type alkaloids, variecolorin A–L, from halotolerant Aspergillus variecolor. Journal of Natural Products. 2007;70:1558–1564. doi: 10.1021/np070208z. [DOI] [PubMed] [Google Scholar]
- Wang W.L., Sun W., Gu Q.Q. 1,6-dihydroxy-3-hydroxtmethyl-8-methoxyanthracene-9,10-dione monohydrate. Acta Crystallographica Section E. 2008;64:o332. doi: 10.1107/S1600536807066986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.L., Zhu T.J., Tao H.W. Three novel, structurally unique spirocyclic alkaloids from the halotolerant B-17 fungal strain of Aspergillus variecolor. Chemistry & Biodiversity. 2007;4:2913–2919. doi: 10.1002/cbdv.200790240. [DOI] [PubMed] [Google Scholar]
- Wang W.L., Zhu T.J., Tao H.W. Two new cytotoxic quinone type compounds from the halotolerant fungus Aspergillus variecolor. The Journal of Antibiotics. 2007;60:603–607. doi: 10.1038/ja.2007.77. [DOI] [PubMed] [Google Scholar]
- Wang W.S., Li X.M., Teuscher F. Chaetopyranin, a benzaldehyde derivative, and other related metabolites from Chaetomium globosum, an endophytic fungus derived from the marine red alga Polysiphonia urceolata. Journal of Natural Products. 2006;69:1622–1625. doi: 10.1021/np060248n. [DOI] [PubMed] [Google Scholar]
- Wang X.N., Radwan M.M., Tarawneh A.H. Antifungal activity against plant pathogens as metabolites from the endophytic fungus Cladosporium cladosporioides. Journal of Agricultural and Food Chemistry. 2013;61:4551–4555. doi: 10.1021/jf400212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J.M., Cole R.J., Kirksey J.W. Emodin, a toxic metabolite of Aspergillus wentii isolated from weevil-damaged chestnuts. Applied Microbiology. 1975;30:26–28. doi: 10.1128/am.30.1.26-28.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Q.Y. Identification of the main species in Fuzhuan Brick Tea. China Tea. 1990;6:2–3. [Google Scholar]
- White T.J., Bruns T., Lee S. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Shinsky T.J., White T.J., editors. PCR protocols: a guide to methods and applications. Academic Press Inc.; New York: 1990. pp. 315–322. [Google Scholar]
- Wilkinson S., Spilsbury J.F. Gliotoxin from Aspergillus chevalieri (Mangin) Thom et Church. Nature. 1965;206:619. doi: 10.1038/206619a0. [DOI] [PubMed] [Google Scholar]
- Williams K., Szwalbe A.J., Mulholland N.P. Heterologous production of fungal maleidrides reveals the cryptic cyclization involved in their biosynthesis. Angewandte Chemie International Edition. 2016;55:6784–6788. doi: 10.1002/anie.201511882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.D., Cheng M.J., Hsieh S.Y. Chemical constituents of the fungus Eurotium chevalieri BCRC 07F0022. Chemistry of Natural Compounds. 2013;49:1175–1176. [Google Scholar]
- Xu A., Wang Y., Wen J. Fungal community associated with fermentation and storage of Fuzhuan brick-tea. International Journal of Food Microbiology. 2011;146:14–22. doi: 10.1016/j.ijfoodmicro.2011.01.024. [DOI] [PubMed] [Google Scholar]
- Yan H.J., Li X.M., Li C.S. Alkaloid and anthraquinone derivatives produced by the marine-derived endophytic fungus Eurotium rubrum. Helvetica Chimica Acta. 2012;95:163–167. [Google Scholar]
- Zalar P., Frisvad J.C., Gunde-Cimerman N. Four new species of Emericella from the Mediterranean region of Europe. Mycologia. 2008;100:779–795. doi: 10.3852/08-078. [DOI] [PubMed] [Google Scholar]
- Zhou L.N., Zhu T.J., Cai S.X. Three new indole-containing diketopiperazine alkaloids from a deep-ocean sediment derived fungus Penicillium griseofulvum. Helvetica Chimica Acta. 2010;93:1758–1762. [Google Scholar]
- Zou X., Li Y., Zhang X., Li Q. A new prenylated indole diketopiperazine alkaloid from Eurotium cristatum. Molecules. 2014;19:17839–17847. doi: 10.3390/molecules191117839. [DOI] [PMC free article] [PubMed] [Google Scholar]