Abstract
We report a novel molecular assay, based on helicase-dependent amplification (HDA), for the detection of enterococci as markers for fecal pollution in water. This isothermal assay targets the same Enterococcus 23S rRNA gene region as the existing quantitative polymerase chain reaction (qPCR) assays of U.S. Environmental Protection Agency Methods 1611 and 1609 but can be entirely performed on a simple heating block. The developed Enterococcus HDA assay successfully discriminated 15 enterococcal from 15 non-enterococcal reference strains and reliably detected 48 environmental isolates of enterococci. The limit of detection was 25 target copies per reaction, only 3 times higher than that of qPCR. The applicability of the assay was tested on 30 environmental water sample DNA extracts, simulating a gradient of fecal pollution. Despite the isothermal nature of the reaction, the HDA results were consistent with those of the qPCR reference. Given this performance, we conclude that the developed Enterococcus HDA assay has great potential as a qualitative molecular screening method for resource-limited settings when combined with compatible up- and downstream processes. This amplification strategy can pave the way for developing a new generation of rapid, low-cost, and field-deployable molecular diagnostic tools for water quality monitoring.
Introduction
For more than a century, the microbiological status of water resources has been assessed on the basis of selective cultivation of fecal indicator bacteria such as Escherichia coli or intestinal enterococci. This approach is well established, has been widely adopted, and represents the methodological and regulatory basis for water quality monitoring in many countries.1,2 However, there are well-known drawbacks and shortcomings associated with it, notably the speed with which results can be provided. Standard culture-based methods for fecal indicator enumeration require 18–72 h.3,4 However, several studies have shown that temporal changes in fecal indicator bacteria levels occur much more rapidly.5−8 As a result, many people may have been exposed to contaminated waters by the time a routine test reveals that there is a problem.
Therefore, an increased level of interest has been directed in the past decade toward the use of molecular methods as rapid detection tools that directly quantify fecal indicator bacterial cells or their intracellular molecules, such as adenosine triphosphate (ATP) or genetic markers (DNA and RNA). Among the explored technologies are flow cytometry,9 immunomagnetic separation/ATP,10,11 and quantitative polymerase chain reaction (qPCR).12,13 Specifically, qPCR has received much attention from researchers and regulators. For the enumeration of enterococci, results of qPCR have been found to correlate with those of traditional culture-based methods13−15 but can be obtained in as few as 3–4 h. This allows a more timely notification of water quality and same-day health warnings. Supported by epidemiological studies,16−18 qPCR was adopted by the U.S. Environmental Protection Agency (U.S. EPA) in 2012 as a recommended method for rapidly enumerating Enterococcus spp. in recreational surface water.19 The two published qPCR-based analysis methods, U.S. EPA Method 161120 and U.S. EPA Method 1609,21 both target a specific 93 bp region of the Enterococcus 23S rRNA gene and can be used by routine laboratories as an option for monitoring recreational beach water quality. However, the implementation of qPCR methods at a point of use can be a challenge. From a methodological standpoint, qPCR requires costly high-end instruments (qPCR thermocycler machines) and extensively trained personnel to perform the method, as well as to analyze and interpret the obtained data. These factors greatly restrict the method’s accessibility and adoption, especially in resource-limited settings.
Isothermal amplification methods, such as helicase-dependent amplification (HDA), address these limitations and offer the opportunity to deliver the benefits of molecular assays beyond well-funded, centralized laboratories. Initially described in 2004,22 HDA is an in vitro DNA amplification method that can be performed at a constant temperature (∼65 °C). Unlike the rapid and repeated heating steps required in PCR, HDA uses the natural unwinding activity of DNA helicase enzymes to separate DNA double strands for subsequent primer annealing and extension. This reaction mechanism facilitates a >1 million-fold amplification of the target sequence without the need for thermal cycling throughout the reaction.22 Thus, HDA can be performed on a simple heating block, in a water bath, or even entirely without electricity (using exothermic chemical heating).23,24 At the same time, HDA offers the advantages of PCR technology, such as high specificity and sensitivity, as well as the capability for multiplex detection.25,26 Furthermore, the relative simplicity of the reaction makes HDA an appealing method for incorporation into portable, battery-operated microfluidic lab-on-the-chip systems.27−30 For instance, HDA-employing microfluidic devices that integrate nucleic acid extraction, amplification, and detection steps in a single device have been described.30,31 These accomplishments highlight the potential of the technique to be used in resource-limited settings by non-experts. Until now, however, HDA has been mainly used for pathogen detection in clinical diagnostics.26,32−35
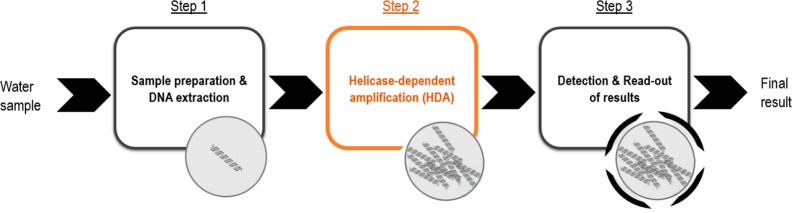
Our vision is to take advantage of HDA amplification to provide a foundation for future low-cost and rapid water quality monitoring tools (Figure 1). The basic premise is nevertheless that the analytical performance of this amplification strategy is shown to be comparable with the qPCR reference. Thus, the aim of this study was to design and develop an HDA assay that is complementary to an existing qPCR assay for the molecular detection of enterococci in environmental waters and to assess its performance as a potential screening tool. The term “complementary” in this context refers to equivalence in terms of specificity and sensitivity (not on being quantitative). To that end, the primers of the qPCR assay of U.S. EPA Method 1611, targeting the 23S rRNA gene as a marker for fecal pollution, were transferred into the HDA reaction format. The performance of the developed Enterococcus HDA assay was evaluated and compared to that of the qPCR reference with respect to specificity, sensitivity, limit of detection, and analysis of environmental isolates, as well as its applicability to environmental water sample DNA extracts.
Figure 1.
Visionary concept of an analytical procedure for water quality testing in which HDA amplification is used for the molecular detection of enterococci in water. Step 1 consists of sample preparation and DNA extraction. Step 2 consists of HDA amplification (focus of this study, development and evaluation). Step 3 consists of detection and readout of results.
Experimental Section
Bacterial Strains
A set of 15 enterococcal (“target strains”) and 15 non-enterococcal reference strains (“nontarget strains”) were used as positive and negative controls, respectively. The target strains were Enterococcus asini (DSM-11492), Enterococcus avium (DSM-20679), Enterococcus casseliflavus (DSM-20680), Enterococcus cecorum (DSM-20682), Enterococcus durans (DSM-20633), Enterococcus faecium (DSM-20477), Enterococcus faecalis (DSM-20478), Enterococcus gallinarum (DSM-20628), Enterococcus hirae (DSM-20160), Tetragenococcus solitarius (also known as Enterococcus solitarius, DSM-5634), Enterococcus sulfureus (DSM-6905), Enterococcus columbae (DSM-7374), Enterococcus mundtii (DSM-4838), Enterococcus dispar (DSM-6630), and Enterococcus raffinosus (DSM-5633). The nontarget strains were Lactococcus garvieae (DSM-6783), Lactococcus lactis subsp. lactis (DSM-20481), Staphylococcus aureus subsp. aureus (DSM-20232), Streptococcus bovis (DSM-20480), Streptococcus salivarius subsp. salivarius (DSM-20560), Tetragenococcus halophilus subsp. halophilus (DSM-20339), Vagococcus fluvialis (DSM-5731), Melissococcus plutonius (DSM-29964), Serratia marcescens subsp. marcescens (DSM-30121), Citrobacter freundii (DSM-30039), Bacillus subtilis (ATCC 6633), E. coli (NCTC 9001), Klebsiella aerogenes (NCTC 9528), Providencia rettgeri (NCTC 7475), and Pseudomonas aeruginosa (NCTC 10662).
Additionally, 45 environmental isolates of enterococci collected from four sampling locations in Austrian surface water bodies throughout the past four seasons were used for the evaluation of the HDA assay. These isolates were cultivated according to the ISO standard.3 Single colonies were picked from culture plates for subsequent DNA extraction and stored at −80 °C. The identities of the environmental enterococcal strains were confirmed using 16S rRNA gene sequencing analysis, as described previously by Ryzinska-Paier et al.36
Environmental Water Samples
A set of 30 environmental water sample DNA extracts were compiled from previous studies, comprising spring/groundwater, surface water, and wastewater samples. Groundwater and spring water samples were taken from the work of Kirschner et al.,37 whereas all surface water samples originated from the Joint Danube Survey 2013.38 Wastewater influent and effluent samples were recovered from various Austrian wastewater treatment plants, described previously by Mayer et al.39
DNA Extraction
Genomic DNA from pure cultures of bacterial reference strains and environmental isolates of enterococci was extracted using the peqGOLD Bacterial DNA Kit (Peqlab, Erlangen, Germany), according to the manufacturer’s protocol. DNA from environmental water samples was extracted from 0.2 μm polycarbonate filters using bead beating and phenol/chloroform as described previously.40 DNA concentrations were measured using the QuantiFluor dsDNA System (Promega, Mannheim, Germany). DNA extracts were stored at −20 and −80 °C until they were further processed with HDA and qPCR in parallel.
Primer and Probe Sequences
All oligonucleotides were synthesized by Eurofins MWG (Ebersberg, Germany). qPCR primer and probe sequences were those described in U.S. EPA Method 161120 and U.S. EPA Method 1609:21 forward primer (5′-GAGAAATTCCAAACGAACTTG), reverse primer (5′-CAGTGCTCTACCTCCATCATT) and TaqMan probe ([6-FAM]-5′-TGGTTCTCTCCGAAATAGCTTTAGGGCTA-TAMRA). For HDA, the same forward and reverse primers were used.
Enterococcus HDA Assay Protocol
Enterococcus HDA reactions were performed on a heating block (PocketBloc, Biozym, Germany), using the IsoAmp II Universal tHDA kit (New England BioLabs, Ipswich, MA) in a final reaction volume of 20 μL. Following a two-step protocol, 10 μL of mix A containing 1× annealing buffer II, each primer at 0.6 μM, and 2.5 μL of DNA was overlaid with mineral oil and heated at 95 °C for 3 min for initial target DNA denaturation. After subsequent equilibration at 65 °C for 3 min, 10 μL of mix B containing 1× annealing buffer II, 8 mM MgSO4, 80 mM NaCl, 3.5 μL of an IsoAmp dNTP solution, and 3.5 μL of IsoAmp enzyme mix was added. Reaction mixtures were then incubated at 65 °C for 90 min. HDA products (10 μL aliquots) were analyzed in 2.5% agarose gels stained with SYBR Gold (Thermo Fisher Scientific, Waltham, MA). HDA reactions that showed a band at the correct size (93 bp) were counted as positive. Unless otherwise noted, amplification reactions were performed in triplicate, including the no-template controls in each run that served as a contamination check.
Enterococcus qPCR Assay Protocol
The Enterococcus qPCR assay was performed in a total reaction volume of 15 μL, containing 1× Kapa Probe Fast (Peqlab), each primer at 1 μM, 80 nM FAM-labeled Enterococcus TaqMan probe, and 2.5 μL of the DNA template. The amplification reactions were performed on a 7500 Fast Real-Time PCR system (Applied Biosystems, New York, NY), according to the following thermal cycling conditions: initial step of 5 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 60 s at 60 °C. Unless otherwise noted, amplification reactions were performed in triplicate, including no-template controls in each run to check for contamination. Quantification was achieved by running a dilution series of plasmid DNA containing the diagnostic fragment of the En. faecalis 23S rRNA gene.
Statistical Analyses
A 95% limit of detection (LOD95%) was defined as that concentration at which a detection probability of 95% is expected. Detection probabilities were modeled as a function of concentration using a logistic regression model, and R software (R Development Core Team, 2008) was employed for this computation. Contingency analysis was conducted in two-by-two contingency tables and by performing χ2 tests, using IBM SPSS Statistics 24 software. For this purpose, triplicates of the HDA and qPCR analyses were clustered: zero positives of three replicates was rated as a negative result, whereas one, two, or three positives of three replicates was rated as a positive result.
Results and Discussion
HDA Assay Development
In an effort to develop an Enterococcus HDA assay, the primer sequences described in the qPCR assay of U.S. EPA Method 161120 were initially assessed in silico using OligoAnalyzer version 3.1 (https://eu.idtdna.com/calc/analyzer) and the Oligonucleotide Properties Calculator (http://www.basic.northwestern.edu/biotools/oligocalc.html). It is specifically relevant for HDA that parameter settings, such as the size, GC content, and melting temperature of the primers, as well as the size and melting temperature of the amplicon, match the criteria of the HDA reaction.41 To experimentally investigate whether the primer set yields an amplification product of the expected size, a subset of bacterial target strains was tested and used to optimize the HDA assay protocol. A series of experiments was performed in which the reaction temperature (60–65 °C) and the concentrations of primers (0.1–1 μM), MgSO4 (4–10 mM), and enzyme mix (2–3-fold excess) were varied to ensure efficient amplification. Final reaction conditions can be found in the Experimental Section.
Sensitivity and Specificity
The sensitivity and specificity of the HDA assay were evaluated with a set of 15 enterococcal (target strains) and 15 non-enterococcal (nontarget) reference strains. These bacterial strains were selected on the basis of the following rationale. Target strains comprised Enterococcus species predominant in human and animal hosts,42 whereas selected nontarget strains included species that are relevant in (fecally polluted) environmental waters and also species that are phylogenetically closely related to enterococci.43 Each strain was analyzed with HDA and qPCR at three different concentrations (1 ng, 10 pg, and 0.1 pg of DNA per reaction) to simulate high, medium, and low bacterial loads, respectively. Table 1 summarizes the results obtained from both methods, including the calculated sensitivity and specificity percentages at each DNA concentration level.
Table 1. Sensitivities and Specificities of HDA and qPCR Evaluated on 15 Enterococcal (target strains) and 15 Non-Enterococcal (nontarget) Reference Strains, Each Tested at Three Different Concentrations (1 ng, 10 pg, and 0.1 pg of DNA per reaction)a.
| HDA |
qPCRf |
|||||
|---|---|---|---|---|---|---|
| 1 ng | 10 pg | 0.1 pg | 1 ng | 10 pg | 0.1 pg | |
| Target Strains | ||||||
| En. asini | 3/3 | 3/3 | 2/3 | 3/3 | 3/3 | 3/3 |
| En. avium | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| En. casseliflavus | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| En. cecorum | 3/3 | 3/3 | 2/3 | 3/3 | 3/3 | 3/3 |
| En. durans | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| En. faecium | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| En. faecalisb | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| En. hirae | 3/3 | 3/3 | 1/3 | 3/3 | 3/3 | 3/3 |
| En. gallinarum | 3/3 | 3/3 | 2/3 | 3/3 | 3/3 | 3/3 |
| T. solitariusc | 3/3 | 3/3 | 2/3 | 3/3 | 3/3 | 3/3 |
| En. sulfureus | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| En. columbae | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| En. mundtii | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| En. dispar | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| En. raffinosus | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| Sensitivityd | 100% | 100% | 87% | 100% | 100% | 100% |
| Nontarget Strains | ||||||
| L. garvieae | 0/3 | 0/3 | 0/3 | 3/3 | 0/3 | 1/3 |
| L. lactis | 2/3 | 0/3 | 0/3 | 3/3 | 0/3 | 0/3 |
| S. aureus | 3/3 | 0/3 | 0/3 | 3/3 | 1/3 | 1/3 |
| St. bovis | 0/3 | 0/3 | 0/3 | 3/3 | 1/3 | 0/3 |
| St. salivarius | 0/3 | 0/3 | 0/3 | 3/3 | 1/3 | 0/3 |
| T. halophilus | 3/3 | 3/3 | 0/3 | 3/3 | 3/3 | 3/3 |
| V. fluvialis | 3/3 | 0/3 | 0/3 | 3/3 | 3/3 | 3/3 |
| M. plutonius | 3/3 | 0/3 | 0/3 | 3/3 | 3/3 | 3/3 |
| Se. marcescens | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
| C. freundii | 3/3 | 1/3 | 0/3 | 3/3 | 1/3 | 0/3 |
| B. subtilis | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
| E. coli | 2/3 | 0/3 | 0/3 | 3/3 | 0/3 | 0/3 |
| K. aerogenes | 0/3 | 0/3 | 0/3 | 2/3 | 0/3 | 0/3 |
| P. rettgeri | 0/3 | 0/3 | 0/3 | 2/3 | 0/3 | 0/3 |
| Ps. aeruginosa | 0/3 | 0/3 | 0/3 | 1/3 | 0/3 | 0/3 |
| Specificitye | 58% | 91% | 100% | 22% | 71% | 76% |
The results are given as the number of positive reactions from triplicate analysis and in the calculated sensitivity and specificity percentages. Results of qPCR were scored negative when the ct value was undetermined or <1 copy was detected.
The analyzed concentrations of 1 ng, 10 pg, and 0.1 pg of DNA are equivalent to approximately 106, 104, and 102 DNA target copies per reaction, respectively, for En. faecalis, with a genome size of 3.22 million bp.55 These DNA copy numbers correspond to 2.5 × 105, 2.5 × 103, and 2.5 × 10 En. faecalis cells per reaction, respectively, considering that, in this species, there are four 23S rRNA gene copies per genome.56 Because the genome size and the 23S rRNA operon number are not known for all species used in this study, DNA concentrations are reported in nanograms rather than in genome equivalents.
Also known as En. solitarius.
Assay sensitivity (%) = 100 × (true positives)/(true positives + false negatives).
Assay specificity (%) = 100 × (true negatives)/(true negatives + false positives).
qPCR data have also been used as a cross-comparison by Martzy et al.57
With respect to sensitivity, the HDA and qPCR results were almost identical. At high (1 ng per reaction) and medium (10 pg per reaction) DNA concentrations, the developed HDA assay reliably detects all Enterococcus strains (100%). A slightly reduced sensitivity (87%) for several Enterococcus species was observed only at low DNA concentrations (0.1 pg per reaction). However, each of those strains gave one or two positive results per triplicate analysis. It must be emphasized that during a screening approach, triplicate results would be clustered; a sample giving at least one positive result would be suspicious and thus would be subject to further testing with qPCR methods. The ability to detect all tested Enterococcus strains highlights HDA’s potential for complementary applications.
In contrast, the specificity of the HDA assay at all tested DNA concentration levels was higher than that of qPCR. However, several nontarget species such as T. halophilus, V. fluvialis, and M. plutonius were codetected via HDA (at high concentrations) and qPCR (even at low concentrations). This can be attributed to the high degree of sequence similarity in the 23S rRNA gene target region between these strains and Enterococcus spp. For instance, M. plutonius (GenBank accession number AJ295317) differs in four single bases from En. faecalis (GenBank accession number AJ295306), and only one of these mutations is located in the primer region (close to the 3′ end of the reverse primer). Surprisingly, despite using the same primers as qPCR, HDA is capable of discriminating between M. plutonius and En. faecalis at medium and low concentrations (zero of three replicates, compared to qPCR that still reports three of three at these levels). We therefore tested whether qPCR achieves similar results when the annealing temperature is increased from 60 to 65 °C, which was the temperature used for the HDA reaction. This modification failed to reduce the number of qPCR false positives (data not shown).
However, from a practical point of view, there are several aspects to consider. First, some of the nontarget strains (phylogenetically related species) have little relevance in environmental waters. Second, the abundance of the nontarget strains at high concentrations (1 ng of DNA of a bacterial nontarget strain per reaction equals ∼105 DNA molecules per reaction, assuming a genome size of approximately 2 million bp) represents a highly unrealistic scenario, as these species represent only a small proportion of microorganisms that might occur in fecally polluted water. Third, false positive results overestimate, rather than underestimate, the presence of enterococci and thus the potential risk of fecally contaminated waters. For a screening method, avoiding false negatives is more important than avoiding false positives. Nevertheless, the developed HDA assay has been shown to specifically discriminate between enterococcal and non-enterococcal strains better than qPCR, at all tested DNA concentration levels, despite the use of the same primers and a predetermined 23S rRNA gene target region.
Limit of Detection
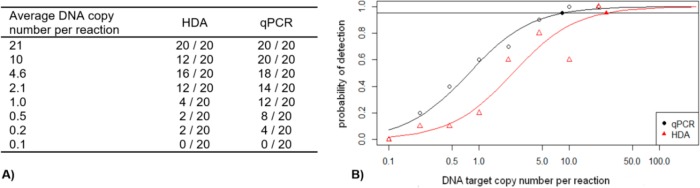
To determine a limit of detection (LOD), a dilution series of genomic DNA of En. faecalis DSM-20478, ranging from 21 to 0.1 target molecules per reaction (with regard to the 23S rRNA gene copies), was analyzed with HDA and qPCR in 20 replicates each. We defined the LOD as the concentration that is detected in at least 95% of the replicates (LOD95%). For this reason, the results of both methods were statistically analyzed with the R software using the logistic regression model (Figure 2). We determined an LOD95% of 25.0 DNA target copies per reaction for the HDA assay and 7.5 DNA target copies per reaction for the reference qPCR assay. Considering the difference in instrument complexity between the two methods and the fact that the primers used for the development of this Enterococcus HDA assay were originally designed for qPCR application, an LOD95% that is only 3 times higher than that of qPCR is surprisingly low and suggests efficient optimization of the HDA reaction conditions.
Figure 2.
Limits of detection (LOD95%). (A) Raw data from the analysis of a dilution series of En. faecalis genomic DNA that served as the input for statistical calculations. (B) Logistic regression model used to determine the LOD95%, which is indicated by filled symbols on the horizontal line.
Analysis of Enterococcus Isolates
To investigate whether the developed HDA assay is capable of detecting environmentally adapted Enterococcus strains, 48 isolates of enterococci from Austrian surface water bodies, cultivated on Slanetz-Bartley and subsequently on Bile Esculin medium,3 were tested with HDA and qPCR at a concentration of 10 pg of enterococcal genomic DNA per reaction. On the basis of 16S rRNA sequencing, these strains were identified as En. faecium (n = 15), En. faecalis (n = 8), En. mundtii (n = 7), En. hirae (n = 7), En. casseliflavus (n = 6), and En. gallinarum (n = 5). In accordance with the qPCR results, the developed HDA assay reliably detects all Enterococcus isolates tested.
Analysis of Environmental Water Sample DNA Extracts
To evaluate the applicability of the developed assay, a total of 30 DNA extracts from environmental water samples were tested with HDA and qPCR in parallel. The sample set consisted of spring/groundwater, surface water, and wastewater (corresponding influents and effluents of wastewater treatment plants), to simulate a range of fecal pollution.
As shown in Table 2, the results were categorized into three classes: (I) unpolluted (spring/groundwater), (II) moderately polluted (surface and polluted spring water), and (III) highly polluted (wastewater) samples. The HDA results agreed with the qPCR results when samples of unpolluted and highly polluted waters were tested. Minor differences between the methods were observed upon analyzing moderately polluted samples. These small variations in triplicate analysis might be attributed to the slightly higher LOD of the HDA. With regard to its use as a potential screening method, however, samples giving at least one positive in a triplicate analysis would be classified positive and thus subjected to further testing with qPCR methods. The high level of agreement of the two methods is also underlined by contingency analysis, which revealed a highly significant and a strong correlation in the performances of HDA and qPCR (χ2 test; ϕ = 1.000, p < 0.001; n = 30).
Table 2. Analysis of a Set of 30 Environmental Water Sample DNA Extracts with and without a History of Fecal Pollutiona.
| DNA extract sample | HDA | qPCRb | mean copy number ± standard deviation determined by qPCRb |
|---|---|---|---|
| Spring/Groundwater Samples (unpolluted) | |||
| GW1 | 0/3 | 0/3 | undetermined |
| GW2 | 0/3 | 0/3 | undetermined |
| GW3 | 0/3 | 0/3 | undetermined |
| GW4 | 0/3 | 0/3 | undetermined |
| GW5 | 0/3 | 0/3 | undetermined |
| SW1 | 0/3 | 0/3 | undetermined |
| SW2 | 0/3 | 0/3 | undetermined |
| SW3 | 0/3 | 0/3 | undetermined |
| SW4 | 0/3 | 0/3 | undetermined |
| SW5 | 0/3 | 0/3 | undetermined |
| Surface Water/Polluted Spring Water Samples (moderately polluted) | |||
| JDS1 | 0/3 | 0/3 | undetermined |
| JDS2 | 1/3 | 3/3 | 6 ± 2 |
| JDS3 | 3/3 | 3/3 | 29 ± 4 |
| JDS4 | 3/3 | 3/3 | 37 ± 9 |
| JDS5 | 2/3 | 3/3 | 19 ± 5 |
| SFW1 | 0/3 | 0/3 | undetermined |
| SFW2 | 3/3 | 3/3 | 156 ± 16 |
| SFW3 | 3/3 | 3/3 | 71 ± 10 |
| SFW4 | 0/3 | 0/3 | undetermined |
| SFW5 | 3/3 | 3/3 | 12 ± 6 |
| Wastewater Samples (highly polluted) | |||
| WWTP1 effluent | 3/3 | 3/3 | 10666 ± 730 |
| WWTP2 effluent | 3/3 | 3/3 | 3369 ± 510 |
| WWTP3 effluent | 3/3 | 3/3 | 1541 ± 172 |
| WWTP4 effluent | 3/3 | 3/3 | 2531 ± 207 |
| WWTP5 effluent | 3/3 | 3/3 | 5803 ± 580 |
| WWTP1 influent | 3/3 | 3/3 | 53085 ± 581 |
| WWTP2 influent | 3/3 | 3/3 | 45690 ± 4073 |
| WWTP3 influent | 3/3 | 3/3 | 41370 ± 1147 |
| WWTP4 influent | 3/3 | 3/3 | 138544 ± 9201 |
| WWTP5 influent | 3/3 | 3/3 | 37803 ± 3354 |
All DNA extracts were measured with HDA and qPCR in parallel. The results of both methods are indicated as the number of positives in triplicate analysis. Results of qPCR were scored negative when the ct value was undetermined or <1 copy was detected. Abbreviations: GW, groundwater; SW, spring water; JDS, Joint Danube Survey; SFW, surface water; WWTP, wastewater treatment plant.
qPCR data have also been used as a cross-comparison by Martzy et al.57
To rule out false-negative results due to inhibition, all samples negative in either qPCR or HDA were additionally tested by being spiked with En. faecalis genomic DNA at a concentration of 100 target copies. Re-analysis of those samples revealed overall positive results, with both methods indicating that no reaction inhibition was taking place, and the original results were indeed truly negative for enterococci.
Implementation of an HDA-Based Screening Method
The developed Enterococcus HDA assay has shown excellent performance, displaying amplification power, analytical specificity, and sensitivity comparable to those of current PCR technology but without the need for expensive equipment in the form of a thermocycler. Likewise, the determined performance characteristics suggest that the developed Enterococcus HDA provides a suitable candidate method for the screening of water samples and a solid basis for developing compatible up- and downstream processes.
As a next step, we plan to develop a simple, user-friendly, and time-saving procedure for sample preparation and DNA extraction (Figure 1, step 1). Our group recently reported methods based on ionic liquids for the direct extraction of DNA from biological materials.44,45 On the basis of this work, we will adapt the procedure for the efficient lysis of enterococcal membrane and cell wall biopolymers to rapidly and quantitatively extract the DNA. The benchmark here will be the sample preparation and DNA extraction procedure of U.S. EPA Methods 1611 and 1609 (DNA extraction using a bead beater after cells were collected on a membrane filter). The efficacy of ionic liquids as cell lysis/DNA extraction solvents was recently shown for Gram-negative bacterial cells (E. coli, ∼88% cell lysis efficiency and Salmonella typhimurium, ∼98% cell lysis efficiency in <5 min).46 Furthermore, we plan to replace the gel electrophoresis method with a rapid (<10 min), inexpensive, and field-deployable nucleic acid lateral flow strip test for the detection of HDA products and the readout of results by the naked eye (Figure 1, step 3). Several detection formats for nucleic acid lateral flow assays are discussed in the literature47 and were reported in combination with HDA by our group48 and others.34,49 In this way, minimally trained personnel can perform the test in settings with poor laboratory infrastructure.
The aforementioned strategies to complement the developed Enterococcus HDA assay can build up a simple and rapid molecular screening method (presence/absence test), which could support making decisions about health risks. For example, in this way, the microbiological status of typically unpolluted water resources such as groundwaters or spring waters can be rapidly assessed in resource-limited settings without sacrificing the advantages of PCR-based methods (i.e., specificity and sensitivity). Only suspicious samples (i.e., positive screening result) would require further testing with sophisticated methods (such as qPCR) to determine the extent of the potential health risk. Thus, such a tool can be beneficial for rationalizing water quality testing by providing an option for rapidly prescreening a large number of samples without delays caused by transporting the samples to centralized laboratories (qPCR methods) or delays caused by the lengthy incubation steps associated with bacterial growth (cultivation-based presence or absence and quantitative tests). Furthermore, it can offer an opportunity for developing countries to gain access to DNA-based water quality testing methods.
Perspectives for the Developed HDA Assay
Miniaturization and integration of the developed Enterococcus HDA assay in microfluidic platforms that are the size of a credit card could offer great opportunities in the future, especially when combined with smartphone technology for optical readout (reviewed in ref (50)). In fact, a variety of microfluidic devices using HDA and other isothermal amplification methods were recently reported as these amplification strategies demand less power and are easier to operate in such devices compared to PCR-based methods.51 Moreover, recent advances in microfabrication have led to the development of rapid, low-cost, and portable microfluidic chip systems for on-site digital isothermal quantification.52−54 For instance, an interesting microfluidic platform − termed SlipChip − was reported that quickly subdivides the reaction mixture into more than thousands of nanoliter-scale compartments with a simple slipping step after pipet loading.53 Robust digital isothermal quantification was achieved by counting the “positive” and “negative” wells using end-fluorescent readout53 or colorimetric readout with an unmodified cell phone camera.54 Integration of the developed Enterococcus HDA assay into such a platform could offer an opportunity to deliver quantitative results and thus an option for monitoring surface waters and recreational bathing areas outside of centralized laboratories by non-experts.
This study aimed to build up the basis for future approaches by providing an isothermal amplification assay that uses the same primers to target the same enterococcal 23S rRNA gene region as used in the existing qPCR methods. This study carefully assessed its applicability as an amplification platform and centerpiece for potential field-deployable tools. Considering the performance of the developed Enterococcus assay, the transfer of helicase-dependent amplification technology into the field of microbiological water quality analysis is encouraging and can offer simple, yet powerful, diagnostic tools for the detection of genetic targets.
Acknowledgments
This study was a joint collaboration of the Interuniversity Cooperation Centre Water & Health (www.waterandhealth.at). This study was part of Life Science Call 2013 Project LSC13-020 funded by the Niederösterreichische Forschungs- and Bildungsgesellschaft (NFB) and Austrian Science Fund (FWF) Project P23900 granted to A.H.F.
The authors declare no competing financial interest.
References
- Guidelines for drinking-water quality; World Health Organization: Geneva, 2011. [Google Scholar]
- Farnleitner A. H.; Ryzinska-Paier G.; Reischer G. H.; Burtscher M. M.; Knetsch S.; Kirschner A. K. T.; Dirnböck T.; Kuschnig G.; Mach R. L.; Sommer R. Escherichia coli and enterococci are sensitive and reliable indicators for human, livestock and wildlife faecal pollution in alpine mountainous water resources. J. Appl. Microbiol. 2010, 109 (5), 1599–1608. 10.1111/j.1365-2672.2010.04788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Water quality – Detection and Enumeration of Intestinal Enterococci – Part 2: Membrane Filtration Method. ISO Technical Report 7899-2:2000; International Organization of Standardization: Geneva, 2000. [Google Scholar]
- Method 1600: Enterococci in water by membran filtration using membrane-enterococcus indoxyl-β-d-glucoside agar (mEI). Technical Report EPA-821-R-09-016; Office of Water, U.S. Environmental Protection Agency: Washington, DC, 2009. [Google Scholar]
- Reischer G. H.; Haider J. M.; Sommer R.; Stadler H.; Keiblinger K. M.; Hornek R.; Zerobin W.; Mach R. L.; Farnleitner A. H. Quantitative microbial faecal source tracking with sampling guided by hydrological catchment dynamics. Environ. Microbiol. 2008, 10 (10), 2598–2608. 10.1111/j.1462-2920.2008.01682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischer G. H.; Kollanur D.; Vierheilig J.; Wehrspaun C.; Mach R. L.; Sommer R.; Stadler H.; Farnleitner A. H. Hypothesis-driven approach for the identification of fecal pollution sources in water resources. Environ. Sci. Technol. 2011, 45 (9), 4038–4045. 10.1021/es103659s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A. B.; Grant S. B.; Kim J. H.; Mowbray S. L.; McGee C. D.; Clark C. D.; Foley D. M.; Wellman D. E. Decadal and shorter period variability of surf zone water quality at Huntington Beach, California. Environ. Sci. Technol. 2002, 36 (18), 3885–3892. 10.1021/es020524u. [DOI] [PubMed] [Google Scholar]
- Leecaster M. K.; Weisberg S. B. Effect of sampling frequency on shoreline microbiology assessments. Mar. Pollut. Bull. 2001, 42 (11), 1150–1154. 10.1016/S0025-326X(01)00130-8. [DOI] [PubMed] [Google Scholar]
- Caruso G.; Monticelli L. S.; Caruso R.; Bergamasco A. Development of a fluorescent antibody method for the detection of Enterococcus faecium and its potential for coastal aquatic environment monitoring. Mar. Pollut. Bull. 2008, 56 (2), 318–324. 10.1016/j.marpolbul.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Bushon R. N.; Brady A. M.; Likirdopulos C. A.; Cireddu J. V. Rapid detection of Escherichia coli and enterococci in recreational water using an immunomagnetic separation/adenosine triphosphate technique. J. Appl. Microbiol. 2009, 106 (2), 432–441. 10.1111/j.1365-2672.2008.04011.x. [DOI] [PubMed] [Google Scholar]
- Lee C. M.; Griffith J. F.; Kaiser W.; Jay J. A. Covalently linked immunomagnetic separation/adenosine triphosphate technique (Cov-IMS/ATP) enables rapid, in-field detection and quantification of Escherichia coli and Enterococcus spp. in freshwater and marine environments. J. Appl. Microbiol. 2010, 109 (1), 324–333. 10.1111/j.1365-2672.2009.04660.x. [DOI] [PubMed] [Google Scholar]
- Ludwig W.; Schleifer K.-H. How Quantitative is Quantitative PCR with Respect to Cell Counts?. Syst. Appl. Microbiol. 2000, 23 (4), 556–562. 10.1016/S0723-2020(00)80030-2. [DOI] [PubMed] [Google Scholar]
- Haugland R. A.; Siefring S. C.; Wymer L. J.; Brenner K. P.; Dufour A. P. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 2005, 39 (4), 559–568. 10.1016/j.watres.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Lavender J. S.; Kinzelman J. L. A cross comparison of QPCR to agar-based or defined substrate test methods for the determination of Escherichia coli and enterococci in municipal water quality monitoring programs. Water Res. 2009, 43 (19), 4967–4979. 10.1016/j.watres.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Noble R. T.; Blackwood A. D.; Griffith J. F.; McGee C. D.; Weisberg S. B. Comparison of rapid quantitative PCR-based and conventional culture-based methods for enumeration of Enterococcus spp. and Escherichia coli in recreational waters. Appl. Environ. Microbiol. 2010, 76 (22), 7437–7443. 10.1128/AEM.00651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade T. J.; Calderon R. L.; Sams E.; Beach M.; Brenner K. P.; Williams A. H.; Dufour A. P. Rapidly Measured Indicators of Recreational Water Quality Are Predictive of Swimming-Associated Gastrointestinal Illness. Environ. Health Perspect. 2006, 114 (1), 24–28. 10.1289/ehp.8273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade T. J.; Calderon R. L.; Brenner K. P.; Sams E.; Beach M.; Haugland R.; Wymer L.; Dufour A. P. High sensitivity of children to swimming-associated gastrointestinal illness: results using a rapid assay of recreational water quality. Epidemiology 2008, 19 (3), 375–383. 10.1097/EDE.0b013e318169cc87. [DOI] [PubMed] [Google Scholar]
- Wade T. J.; Sams E.; Brenner K. P.; Haugland R.; Chern E.; Beach M.; Wymer L.; Rankin C. C.; Love D.; Li Q.; Noble R.; Dufour A. P. Rapidly measured indicators of recreational water quality and swimming-associated illness at marine beaches: a prospective cohort study. Environ. Health 2010, 9 (1), 66. 10.1186/1476-069X-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recreational water quality criteria. Technical Report EPA-820-F-12-058; Office of Water, U.S. Environmental Protection Agency: Washington, DC, 2012. [Google Scholar]
- Method 1611: Enterococci in Water by TaqMan quantitative polymerase chain reaction (qPCR) assay. Technical Report EPA-821-R-12-008; Office of Water, U.S. Environmental Protection Agency: Washington, DC, 2012. [Google Scholar]
- Method 1609: Enterococci in Water by TaqMan Quantitative Polymerase Chain Reaction (qPCR) with Internal Amplification Control (IAC) Assay. Technical Report EPA-820-R-13-005; Office of Water, U.S. Environmental Protection Agency: Washington, DC, 2013. [Google Scholar]
- Vincent M.; Xu Y.; Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004, 5 (8), 795. 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.; Do J.; Mahalanabis M.; Fan A.; Zhao L.; Jepeal L.; Singh S. K.; Klapperich C. M. Low cost extraction and isothermal amplification of DNA for infectious diarrhea diagnosis. PLoS One 2013, 8 (3), e60059. 10.1371/journal.pone.0060059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBarre P.; Hawkins K. R.; Gerlach J.; Wilmoth J.; Beddoe A.; Singleton J.; Boyle D.; Weigl B. A simple, inexpensive device for nucleic acid amplification without electricity—toward instrument-free molecular diagnostics in low-resource settings. PLoS One 2011, 6 (5), e19738. 10.1371/journal.pone.0019738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.; Easley C. J. Isothermal DNA amplification in bioanalysis: strategies and applications. Bioanalysis 2011, 3 (2), 227–239. 10.4155/bio.10.172. [DOI] [PubMed] [Google Scholar]
- Doseeva V.; Forbes T.; Wolff J.; Khripin Y.; O’Neil D.; Rothmann T.; Nazarenko I. Multiplex isothermal helicase-dependent amplification assay for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. Diagn. Microbiol. Infect. Dis. 2011, 71 (4), 354–365. 10.1016/j.diagmicrobio.2011.08.021. [DOI] [PubMed] [Google Scholar]
- Asiello P. J.; Baeumner A. J. Miniaturized isothermal nucleic acid amplification, a review. Lab Chip 2011, 11 (8), 1420–1430. 10.1039/c0lc00666a. [DOI] [PubMed] [Google Scholar]
- Andresen D.; Nickisch-Rosenegk M. v.; Bier F. F. Helicase-dependent amplification: use in OnChip amplification and potential for point-of-care diagnostics. Expert Rev. Mol. Diagn. 2009, 9 (7), 645–650. 10.1586/erm.09.46. [DOI] [PubMed] [Google Scholar]
- Kaprou G. D.; Papadakis G.; Papageorgiou D. P.; Kokkoris G.; Papadopoulos V.; Kefala I.; Gizeli E.; Tserepi A. Miniaturized devices for isothermal DNA amplification addressing DNA diagnostics. Microsyst. Technol. 2016, 22, 1529. 10.1007/s00542-015-2750-x. [DOI] [Google Scholar]
- Mahalanabis M.; Do J.; Almuayad H.; Zhang J. Y.; Klapperich C. M. An integrated disposable device for DNA extraction and helicase dependent amplification. Biomed. Microdevices 2010, 12 (2), 353–359. 10.1007/s10544-009-9391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Park S.; Liu K.; Tsuan J.; Yang S.; Wang T.-H. A surface topography assisted droplet manipulation platform for biomarker detection and pathogen identification. Lab Chip 2011, 11 (3), 398–406. 10.1039/C0LC00296H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill P.; Alvandi A.-H.; Abdul-Tehrani H.; Sadeghizadeh M. Colorimetric detection of Helicobacter pylori DNA using isothermal helicase-dependent amplification and gold nanoparticle probes. Diagn. Microbiol. Infect. Dis. 2008, 62 (2), 119–124. 10.1016/j.diagmicrobio.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Torres-Chavolla E.; Alocilja E. C. Nanoparticle based DNA biosensor for tuberculosis detection using thermophilic helicase-dependent isothermal amplification. Biosens. Bioelectron. 2011, 26 (11), 4614–4618. 10.1016/j.bios.2011.04.055. [DOI] [PubMed] [Google Scholar]
- Chow W. H. A.; McCloskey C.; Tong Y.; Hu L.; You Q.; Kelly C. P.; Kong H.; Tang Y. W.; Tang W. Application of isothermal helicase-dependent amplification with a disposable detection device in a simple sensitive stool test for toxigenic Clostridium difficile. J. Mol. Diagn. 2008, 10, 452. 10.2353/jmoldx.2008.080008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresen D.; von Nickisch-Rosenegk M.; Bier F. F. Helicase dependent OnChip-amplification and its use in multiplex pathogen detection. Clin. Chim. Acta 2009, 403 (1), 244–248. 10.1016/j.cca.2009.03.021. [DOI] [PubMed] [Google Scholar]
- Ryzinska-Paier G.; Sommer R.; Haider J. M.; Knetsch S.; Frick C.; Kirschner A. K. T.; Farnleitner A. H. Acid phosphatase test proves superior to standard phenotypic identification procedure for Clostridium perfringens strains isolated from water. J. Microbiol. Methods 2011, 87 (2), 189–194. 10.1016/j.mimet.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner A. K. T.; Kavka G.; Reischer G. H.; Sommer R.; Blaschke A. P.; Stevenson M.; Vierheilig J.; Mach R. L.; Farnleitner A. H.. Microbiological Water Quality of the Danube River: Status Quo and Future Perspectives. In The Danube River Basin; Springer: Dordrecht, The Netherlands, 2014; pp 439–468. [Google Scholar]
- Joint Danube Survey 3. A Comprehensive Analysis of Danube Water Quality; ICPDR-International Commission for the Protection of the Danube River: Vienna, 2015. [Google Scholar]
- Mayer R. E.; Bofill-Mas S.; Egle L.; Reischer G. H.; Schade M.; Fernandez-Cassi X.; Fuchs W.; Mach R. L.; Lindner G.; Kirschner A.; Gaisbauer M.; Piringer H.; Blaschke A. P.; Girones R.; Zessner M.; Sommer R.; Farnleitner A. H. Occurrence of human-associated Bacteroidetes genetic source tracking markers in raw and treated wastewater of municipal and domestic origin and comparison to standard and alternative indicators of faecal pollution. Water Res. 2016, 90, 265–276. 10.1016/j.watres.2015.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R. I.; Whiteley A. S.; O’Donnell A. G.; Bailey M. J. Rapid Method for Coextraction of DNA and RNA from Natural Environments for Analysis of Ribosomal DNA- and rRNA-Based Microbial Community Composition. Appl. Environ. Microbiol. 2000, 66 (12), 5488–5491. 10.1128/AEM.66.12.5488-5491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biohelix IsoAmp II Universal tHDA KIt. http://www.biohelix.com/pdf/H0110S_full_version_BH_NG_eDatacard.pdf.
- Layton B. A.; Walters S. P.; Lam L. H.; Boehm A. B. Enterococcus species distribution among human and animal hosts using multiplex PCR. J. Appl. Microbiol. 2010, 109 (2), 539–547. 10.1111/j.1365-2672.2010.04675.x. [DOI] [PubMed] [Google Scholar]
- Behr T.; Koob C.; Schedl M.; Mehlen A.; Meier H.; Knopp D.; Frahm E.; Obst U.; Schleifer K. H.; Niessner R.; Ludwig W. A nested array of rRNA targeted probes for the detection and identification of enterococci by reverse hybridization. Syst. Appl. Microbiol. 2000, 23 (4), 563–572. 10.1016/S0723-2020(00)80031-4. [DOI] [PubMed] [Google Scholar]
- Gonzalez García E.; Ressmann A. K.; Gaertner P.; Zirbs R.; Mach R. L.; Krska R.; Bica K.; Brunner K. Direct extraction of genomic DNA from maize with aqueous ionic liquid buffer systems for applications in genetically modified organisms analysis. Anal. Bioanal. Chem. 2014, 406 (30), 7773–7784. 10.1007/s00216-014-8204-y. [DOI] [PubMed] [Google Scholar]
- Ressmann A. K.; García E. G.; Khlan D.; Gaertner P.; Mach R. L.; Krska R.; Brunner K.; Bica K. Fast and efficient extraction of DNA from meat and meat derived products using aqueous ionic liquid buffer systems. New J. Chem. 2015, 39 (6), 4994–5002. 10.1039/C5NJ00178A. [DOI] [Google Scholar]
- Fuchs-Telka S.; Fister S.; Mester P.-J.; Wagner M.; Rossmanith P. Hydrophobic ionic liquids for quantitative bacterial cell lysis with subsequent DNA quantification. Anal. Bioanal. Chem. 2017, 409 (6), 1503–1511. 10.1007/s00216-016-0112-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma-Trumpie G. A.; Korf J.; van Amerongen A. Lateral flow (immuno)assay: its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 2009, 393 (2), 569–582. 10.1007/s00216-008-2287-2. [DOI] [PubMed] [Google Scholar]
- Kolm C.; Mach R. L.; Krska R.; Brunner K. A rapid DNA lateral flow test for the detection of transgenic maize by isothermal amplification of the 35S promoter. Anal. Methods 2015, 7 (1), 129–134. 10.1039/C4AY01997K. [DOI] [Google Scholar]
- Kim H.-J.; Tong Y.; Tang W.; Quimson L.; Cope V. A.; Pan X.; Motre A.; Kong R.; Hong J.; Kohn D.; Miller N. S.; Poulter M. D.; Kong H.; Tang Y.-W.; Yen-Lieberman B. A rapid and simple isothermal nucleic acid amplification test for detection of herpes simplex virus types 1 and 2. J. Clin. Virol. 2011, 50 (1), 26–30. 10.1016/j.jcv.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauk M. G.; Liu C.; Sadik M.; Bau H. H.. Microfluidic Devices for Nucleic Acid (NA) Isolation, Isothermal NA Amplification, and Real-Time Detection. In Mobile Health Technologies: Methods and Protocols; Rasooly A., Herold K. E., Eds.; Springer: New York, 2015; pp 15–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tröger V.; Niemann K.; Gärtig C.; Kuhlmeier D. Isothermal Amplification and Quantification of Nucleic Acids and its Use in Microsystems. J. Nanomed. Nanotechnol. 2015, 6 (3), 1. 10.4172/2157-7439.1000282. [DOI] [Google Scholar]
- Yeh E.-C.; Fu C.-C.; Hu L.; Thakur R.; Feng J.; Lee L. P. Self-powered integrated microfluidic point-of-care low-cost enabling (SIMPLE) chip. Sci. Adv. 2017, 3 (3), e1501645. 10.1126/sciadv.1501645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F.; Davydova E. K.; Du W.; Kreutz J. E.; Piepenburg O.; Ismagilov R. F. Digital isothermal quantification of nucleic acids via simultaneous chemical initiation of recombinase polymerase amplification reactions on SlipChip. Anal. Chem. 2011, 83 (9), 3533. 10.1021/ac200247e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Manzano J.; Karymov M. A.; Begolo S.; Selck D. A.; Zhukov D. V.; Jue E.; Ismagilov R. F. Reading Out Single-Molecule Digital RNA and DNA Isothermal Amplification in Nanoliter Volumes with Unmodified Camera Phones. ACS Nano 2016, 10 (3), 3102–3113. 10.1021/acsnano.5b07338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen I. T.; Banerjei L.; Myers G. S. A.; Nelson K. E.; Seshadri R.; Read T. D.; Fouts D. E.; Eisen J. A.; Gill S. R.; Heidelberg J. F.; Tettelin H.; Dodson R. J.; Umayam L.; Brinkac L.; Beanan M.; Daugherty S.; DeBoy R. T.; Durkin S.; Kolonay J.; Madupu R.; Nelson W.; Vamathevan J.; Tran B.; Upton J.; Hansen T.; Shetty J.; Khouri H.; Utterback T.; Radune D.; Ketchum K. A.; Dougherty B. A.; Fraser C. M. Role of Mobile DNA in the Evolution of Vancomycin-Resistant Enterococcus faecalis. Science 2003, 299 (5615), 2071. 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- Marshall S. H.; Donskey C. J.; Hutton-Thomas R.; Salata R. A.; Rice L. B. Gene Dosage and Linezolid Resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 2002, 46 (10), 3334–3336. 10.1128/AAC.46.10.3334-3336.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martzy R.; Kolm C.; Brunner K.; Mach R. L.; Krska R.; Sinkovec H.; Sommer R.; Farnleitner A. H.; Reischer G. H. A loop-mediated isothermal amplification (LAMP) assay for the rapid detection of Enterococcus spp. in water. Water Res. 2017, n/a. 10.1016/j.watres.2017.05.023. [DOI] [PubMed] [Google Scholar]