Fig. 3.
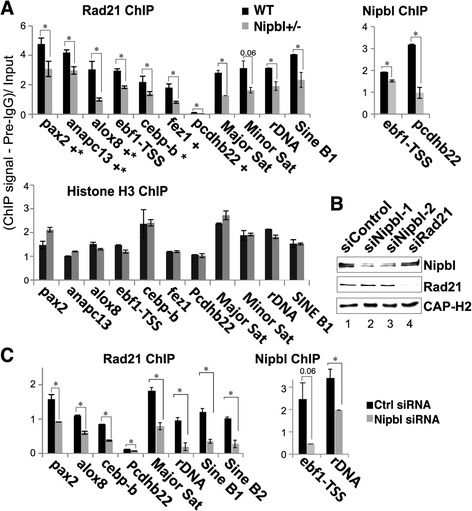
Nipbl reduction decreases cohesin binding. a Manual ChIP-q-PCR of cohesin-binding sites at unique gene regions and repeat regions using anti-Rad21 antibody (top left) compared to histone H3 (bottom) in Nipbl+/− mutant and wild type MEFs. Representative examples of Nipbl ChIP are also shown (top, right). “Plus sign” indicates CTCF binding, and “asterisk” indicates the presence of motif. PCR signals were normalized with preimmune IgG (pre-IgG) and input. *p < 0.05. b Western blot analysis of control, Nipbl, or Rad21 siRNA-treated cells is shown using antibodies indicated. Depletion efficiency and specificity of Nipbl siRNA were also examined by RT-q-PCR (Table 2). Nipbl protein depletion was estimated to be ~ 80% (siNipbl-1) and 60% (siNipbl-2) according to densitometirc measurement (lanes 2 and 3, respectively). Comparable ChIP results were obtained by the two Nipbl siRNAs (data not shown). c Similar manual ChIP-q-PCR analysis as in a in control and Nipbl siRNA (siNipbl-1)-treated MEFs
