Fig. 8.
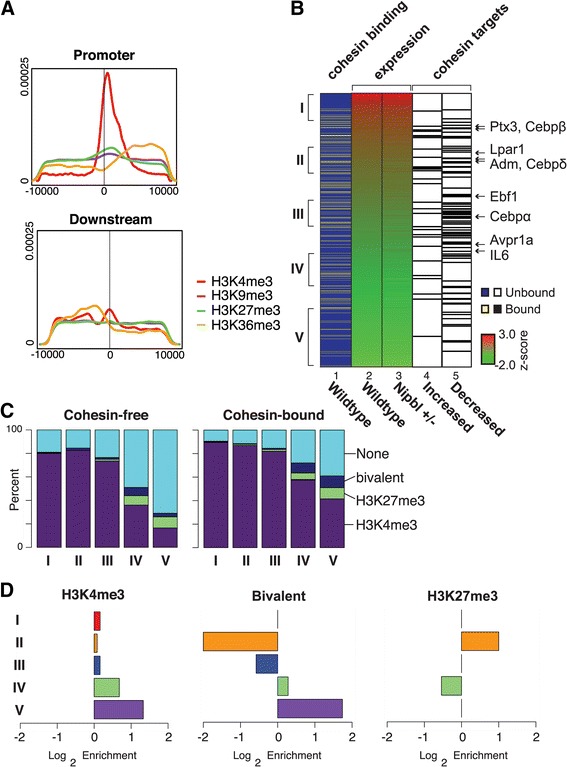
Enrichment of H3K4me3 at the promoters of cohesin-bound genes. a Density of histone modifications within 10 kb of cohesin peaks found in the promoter or downstream regions. Histone methylation data was downloaded from NCBI (GEO: GSE26657). Tags within a 10-kb window around cohesin peaks located in a promoter region were counted and normalized to the total number of tags (reads per million) and used to generate a density plot. b Expression status of cohesin target genes. Genes are ranked by their expression status (shown as a z-score) in wild type MEFs (lane 2), and those genes with cohesin binding at the promoter regions are indicated by yellow lines (lane 1). The expression status of the corresponding genes in Nipbl mutant cells is also shown (lane 3), and the cohesin target genes (Table 2) (either upregulated (lane 4) or downregulated (lane 5) in mutant cells) are indicated by black lines. Genes in the adipogenesis pathway are indicated with arrows on the right. Five clusters (I through V) of 200 cohesin-bound genes each in wild type MEFs according to the expression levels are indicated on the left, which were used for the analysis in c and d. c The numbers of cohesin target genes containing histone marks in the promoter were tallied for the categories I through V from b. As a control, the cohesin-free gene directly below each cohesin target gene was also tallied and plotted. H3K4me3, H3K9me3, H3K27me3, bivalent (H3K4me3 and H3K27me3), and the promoters with none of these marks (“None”) are indicated. There is almost no signal of H3K9me3 in these categories. d Enrichment plot of H3K4me3, H3K27me3, and bivalent (H3K4me3 and K27me3) in promoters of cohesin-bound genes versus cohesin-free genes in the five expression categories as in c is shown
