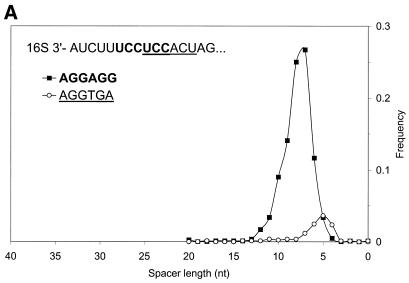
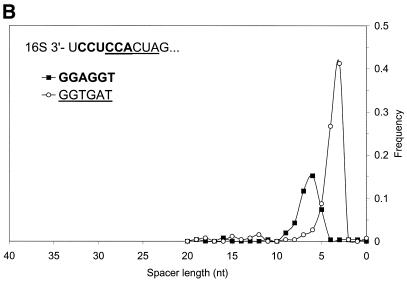
Figure 9.
(A) Distribution of spacer lengths observed in the B.subtilis genome for two different types of possible RBS hexamers: AGGAGG and AGGTGA. Multiple alignment allows these hexamers to be superimposed. In actual upstream sequences, these hexamers tend to occupy different locations relative to the start codon. This preference may be involved in the precise positioning of the ribosome at the translation initiation site when the 16S rRNA binds to mRNA. The more frequent hexamer was observed on average at a further distance from the gene start than the rare hexamer. (B) Distribution of spacer lengths observed in the M.thermoautotrophicum genome for two different types of RBS hexamers: GGAGGT and GGTGAT. Properties of these hexamers are similar to the two hexamers observed in the B.subtilis genome (A), except that more frequent hexamer is now found on average at a closer distance to the gene start than the rare hexamer.