Figure 4.
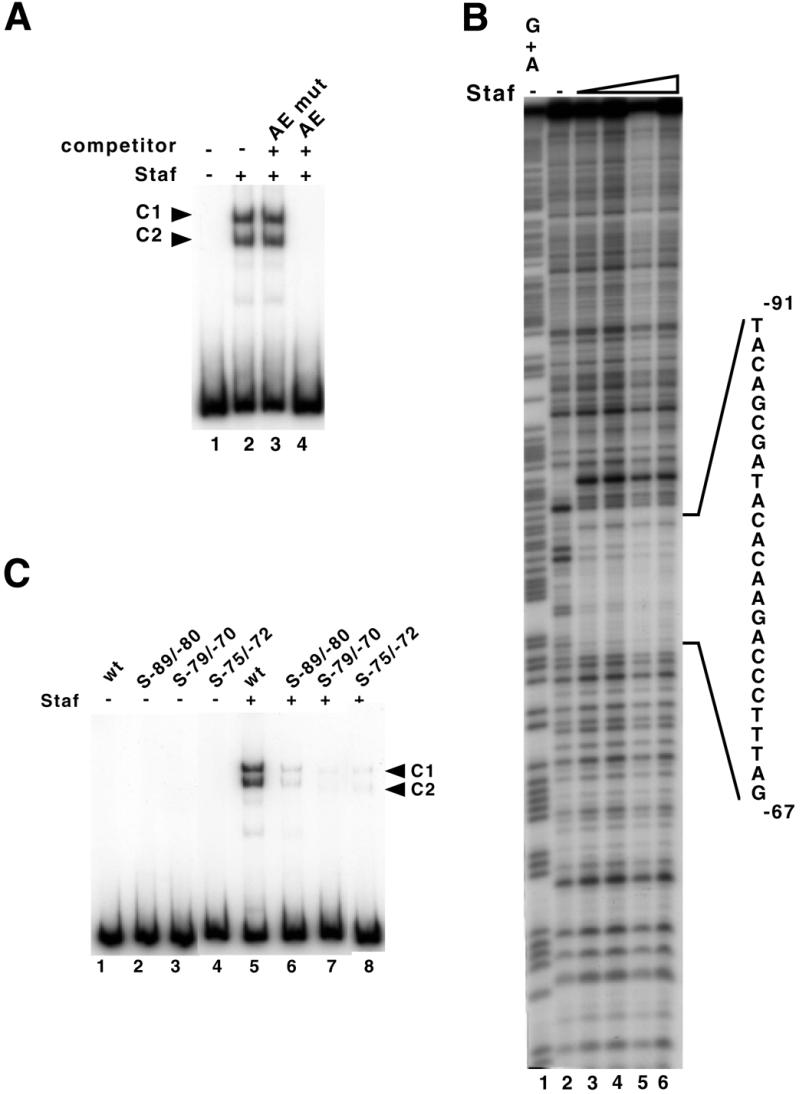
Identification of the Staf binding site in the human H1 RNA gene. (A) Gel retardation assay with a labeled DNA fragment encompassing positions –279/–1 upstream of the transcription start site. The 32P-end-labeled DNA probe was incubated in the absence (lane 1) or presence (lanes 2–4) of Staf-containing bacterial extracts. Reactions in lanes 3 and 4 contained a 1000-fold excess of unlabeled unspecific (AE mut) and specific (AE) competitor, respectively. (B) Footprint analysis of Staf–DNA complexes formed on the H1 RNA gene. DNase I digestion of the human H1 probe, labeled on the template strand, in the absence (lane 2) or presence (lanes 3–6) of increasing amounts of Staf. Probes submitted to G + A chemical cleavage were used as markers (lane 1). The protected region is indicated. (C) Staf binding assays on wild-type and mutant versions of the human H1 RNA gene. A 223 bp end-labeled fragment containing wild-type or mutant versions of the Staf binding site was used in the binding studies. Lanes 1–4, no protein added. Lanes 5–8 contained 1 µl of 20-fold diluted bacterial extracts containing the recombinant Staf. Probes are indicated above the lanes.
