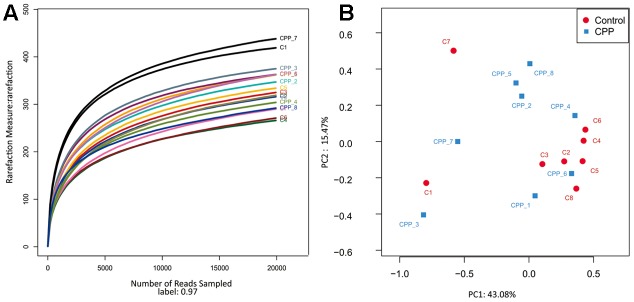
FIGURE 2.
Rarefaction curve of 16S rDNA sequences of the 16 samples and principal coordinate analysis (PCoA) of the samples using Unweighted-UniFrac from pyrosequencing. (A) Rarefaction curves based on the 16S rRNA gene sequencing of the 16 samples from the METH CPP group and the control group (C represents the control group, CPP represents the METH CPP group). The rarefaction curves suggested that the bacterial community was represented well because the curves became relatively flat as the number of sequences analyzed increased. (B) Principal coordinate analysis (PCoA) of the samples using Unweighted-UniFrac from pyrosequencing. The red dots represent the control group, and the blue squares represent the METH-CPP group. The fecal microbiotas of the two groups could not be divided into clusters according to community composition using Unweighted UniFrac metrics and could not be separated clearly by PCoA analysis (ADONIS test, R2 = 0.08276, p-value = 0.191).