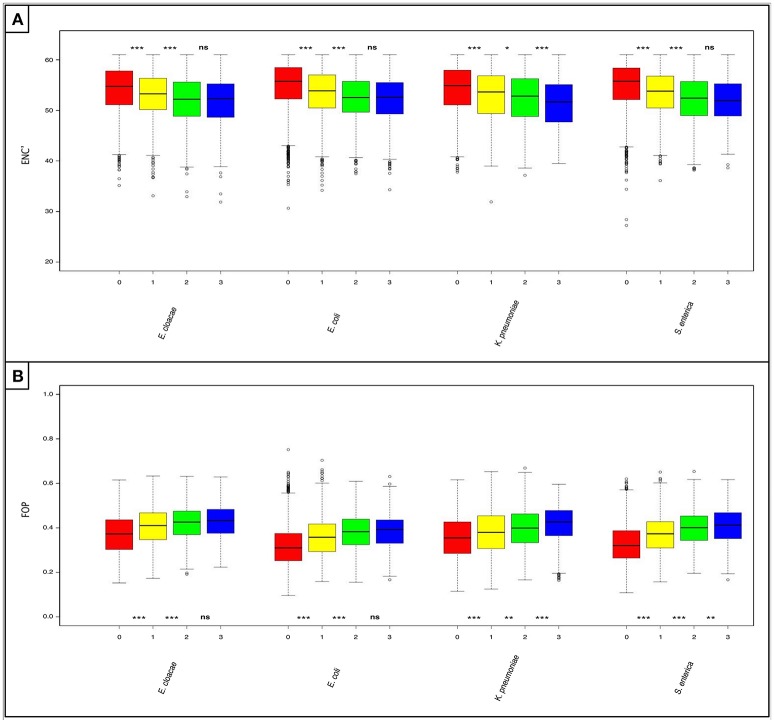
Figure 3.
Horizontally acquired genes that are shared by more of the studied Enterobacteriaceae species are better adapted to the codon usage of their host species. Boxplots depicting: (A) levels of codon usage bias (as measured using ENC') and (B) frequency of the optimal codons (FOP) in the “rare” pangenes. For each species four boxes are given: red—“rare” pangenes that are not shared in any of the three other species; yellow—“rare” pangenes shared by one additional species; green—“rare” pangenes shared by two additional species; blue—“rare” pangenes shared by all species. Whisker length for each boxplot represents 1.5 IQR. Statistical significance of differences between the gene-loss groups according to a non-paired, one-sided Mann-Whitney-Wilcoxon test is denoted by: ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05, and (ns) for P>0.05.