Figure 2.
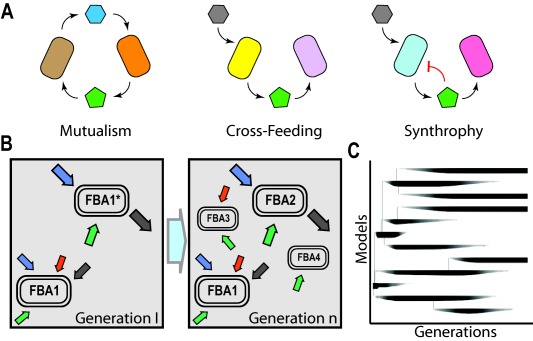
A. Metabolic interactions that can take place in a population. Cells can exchange metabolites that are required to support each other's growth in a mutualistic interaction (left). One of the cells can use a metabolite excreted by another cell, favouring in this way the metabolism of the producer through the pathways leading to the excretion (centre). When the metabolites excreted have an inhibitory effect on the producer (e.g. because they lead to thermodynamic equilibrium), the relationship with a degrader cell of the inhibitory metabolite is mutually beneficial and known as syntrophy (right).
B. Dynamic modelling of the evolution of Flux Balance Analysis models. Cells can be modelled as metabolic networks exchanging metabolites with other cells in the population. In this abstraction each cell is represented by a FBA model. These models can replicate over time and also evolve, producing populations composed by models with different constrains for uptake and secretion of metabolites.
C. Dynamic analysis of model genealogy. The frequency of each model in the population changes over time being the darkest bars the most abundant models. Due to mutations, new models arise and they are represented as new branches in the phylogeny. Plot redrawn from Großkopf et al. (2016).
