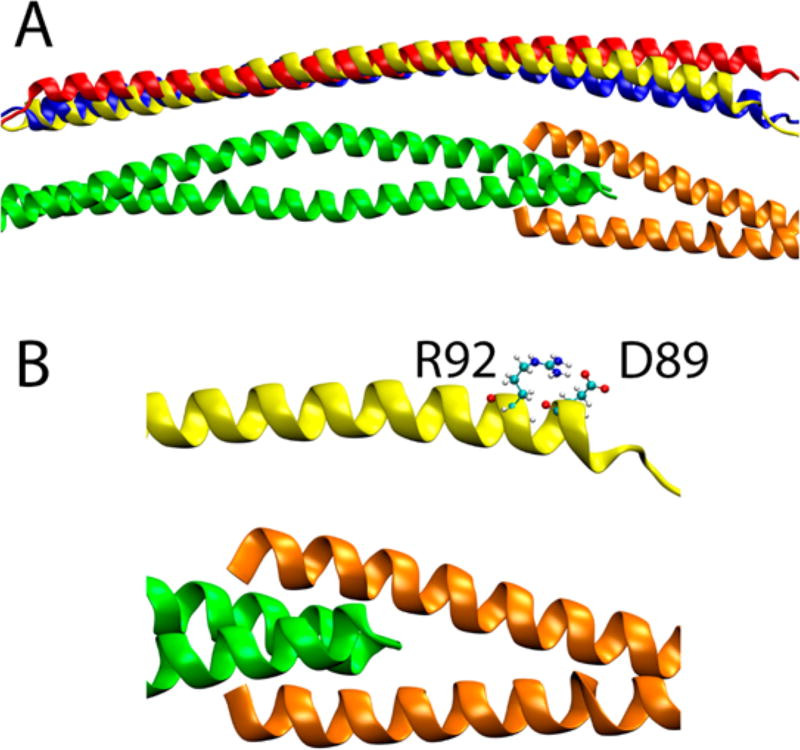
Figure 9.
(A) Average structures of molecular dynamics simulations for wild-type, D230N, and R92L complexes. The Tm’s are shown as the wild type, while the positioning of the cTnT of the mutants is shown with the Tm’s aligned with the wild-type structures. The C-terminal end of Tm is colored green, the N-terminal end of Tm orange, wild-type cTnT yellow, D230N blue, and R92L red. (B) Closer view of the N-terminus of the cTnT. The cTnT D89 and R92 side chains that create the salt bridge that is eliminated by the R92L mutation are shown in ball-and-stick representation. Cyan spheres are carbon atoms, white spheres hydrogen atoms, blue spheres nitrogen atoms, and red spheres oxygen atoms.