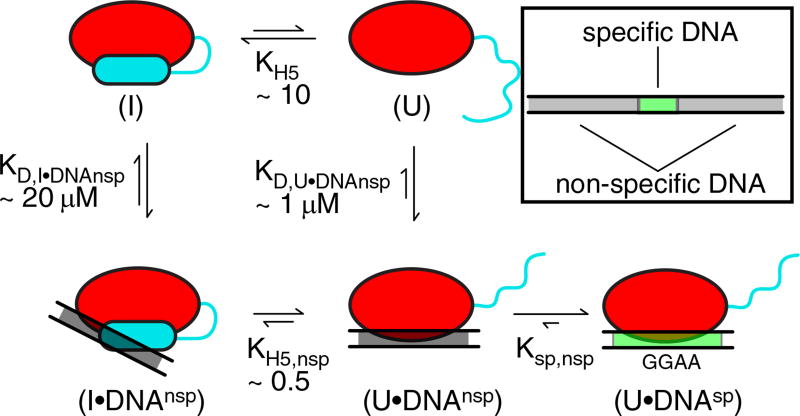
Fig. 8.
Thermodynamic model of ETV6 autoinhibition. The interaction of the ETV6 ETS domain with specific DNA in a background of excess non-specific DNA can be represented by at least a five-state model (red oval, core ETS domain; cyan cylinder or line, folded or unfolded inhibitory helix H5, respectively; gray rectangle, nonspecific DNAnsp; and green rectangle, specific DNAsp). Additional states are possible, but not shown for simplicity. Free ETV6 can adopt inhibited (I) and uninhibited (U) states with helix H5 folded or unfolded, respectively. Inhibited ETV6 interacts with DNAnsp via a displaced DNA-binding interface, whereas the uninhibited ETV6 interacts via the canonical binding interface. Once ETV6 encounters DNAsp, a high-affinity complex is formed. The measured or calculated equilibrium constants for H5 folding in the absence (KH5) or presence (KH5,nsp) of DNAnsp and macroscopic dissociation constants for the protein• DNAnsp complexes with H5 folded (KD,I•DNAnsp) or unfolded (KD,U•DNAnsp) are shown. The macroscopic KD values include binding to multiple sub-sites within the 15-bp duplex DNA used as DNAnsp, of which only one schematic binding mode is shown. Also indicated is the intramolecular dissociation constant from the specific to nonspecific complexes (Ksp,nsp). The clockwise pathway from (I) to (U•DNAnsp) corresponds to a conformational selection model, whereas the counterclockwise pathway can be viewed as an induced fit model.