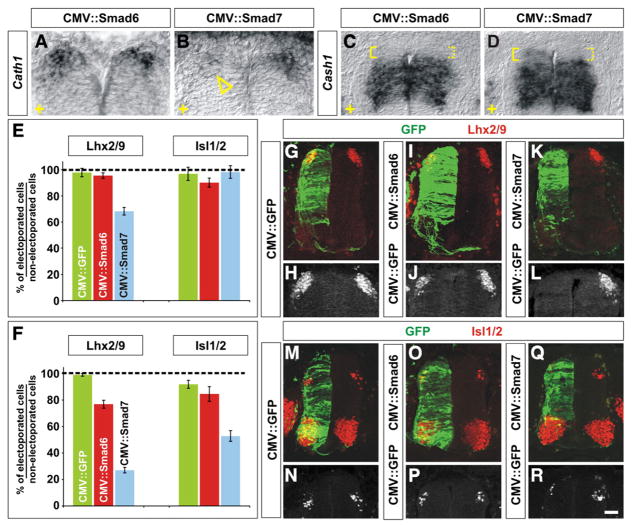
Fig. 2.
Mis-expression of I-Smads differentially affects the fate of dorsal spinal neurons. (A–R) GFP (G, H, M and N), Smad6 (A, C, I, J, O and P) or Smad7 (B, D, K, L, Q and R) were ectopically expressed throughout the spinal cord under the control of the CMV enhancer by in ovo electroporation at either HH stages 10–12 (E) or HH stages 14–16 (A–D, F–R). Embryos were harvested at HH stages 24/25 and examined for the extent of Cath1 (A and B) and Cash1 (C and D) expression and the number of Lhx2/9+ commissural (dI1) neurons (red, G–L) or Isl1/2+ association (dI3) neurons (red, M–R). (A–D) In situ hybridization experiments demonstrated that in ovo electroporation of Smad7 (arrowhead, B), but not Smad6 (A), greatly reduces the extent of expression of Cath1 expression (A and B) suggesting that dP1 cells have been lost. Cash1 levels were also affected by Smad7 (D), but Smad6 (C), mis-expression, low levels of Cash1 expression were now observed adjacent to the roof plate (compare expression levels in solid brackets to dotted brackets, C and D). The electroporated side is indicted by +. (E) The ectopic expression of Smad6 from HH stages 10–12 had no effect on the number of either dI1 or dI3 neurons compared to the electroporation of GFP (dI1, Student’s t-test, p>0.53 probability different from GFP+ control, n=107 sections from 8 embryos; dI3, p>0.29, n=50 sections from 5 embryos). In contrast, the mis-expression of Smad7 resulted in a 30% loss of dI1 neurons (p<4.3×10−11, n=72 sections from 4 embryos) but had no effect on the number of dI3 neurons (p>0.70, n=47 sections from 6 embryos). (F–R) A CMV::GFP construct electroporated into the chick spinal cord at HH stages 14–16 had no observable effect on the number of either Lhx2/9+ dI1 (F, G and H; n=316 sections from 8 embryos) or Isl1/2+ dI3 neurons (F, M and N; n=72 sections from 5 embryos). The mis-expression of Smad6 construct had a weak effect on cell fate: 20% of dI1 neurons were lost (F, I and J; p<6.4×10−17 probability different from GFP+ control, n=69 sections from 5 embryos) but there was no significant effect of mis-expressing Smad6 on the number of dI3 neurons (F, O and P; p>0.74, n=45 sections from 5 embryos). In contrast, the mis-expression of Smad7 resulted in a profound loss of both dI1 and dI3 neurons. Over 70% of dI1 neurons were lost (F, K and L; p<2.7×10−131, n=105 sections from 9 embryos) whereas about 50% of dI3 neurons were lost (F, Q and R; p<2.2×10−11, n=80 sections from 5 embryos) from HH stage 24/25 spinal cords. Scale bar: 25 μm.