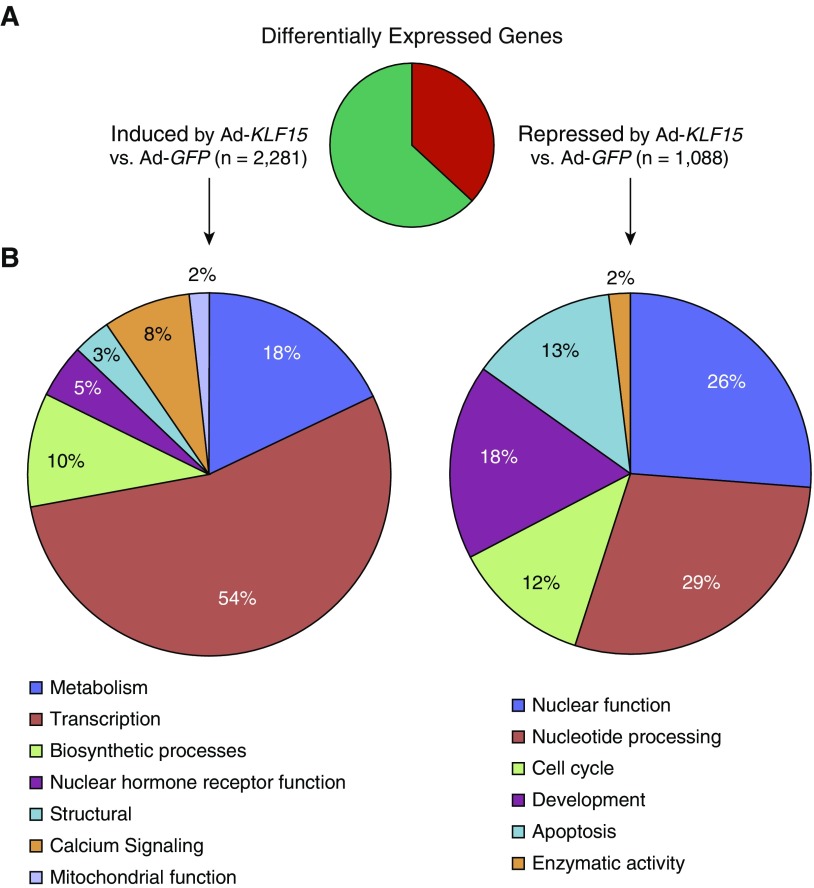
Figure 2.
Global analysis of KLF15-regulated transcription in HASM cells using RNA-seq. (A) Pie chart summarizing differentially expressed genes, defined as those exhibiting twofold or greater change in expression (adjusted P < 0.05) with Ad-KLF15 compared with Ad-GFP and a minimum mean read number of 30. (B) The DAVID Gene Ontology Analysis tool was applied to 200 of the top most strongly regulated transcripts (based on P value) within gene sets significantly induced (left) or repressed (right) by Ad-KLF15. We considered clusters with enrichment scores greater than 1 as significant. Pie charts illustrate distinct categories of significant functional clusters of related gene ontology terms within each gene set.