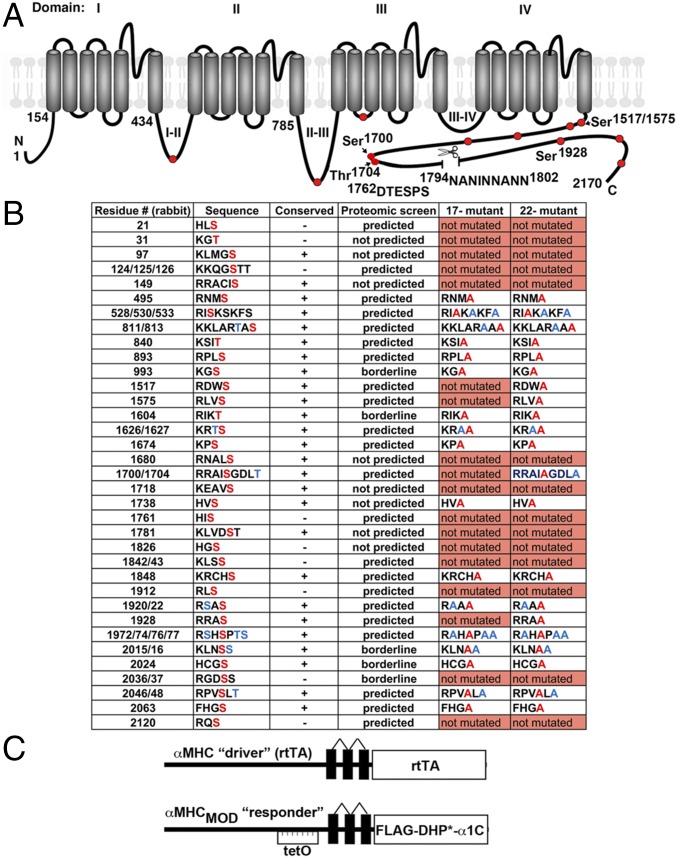
Fig. 1.
Putative PKA phosphorylation sites and proteolytic cleavage site in CaV1.2 α1C. (A) Schematic of rabbit cardiac α1C subunit topology. The putative proteolytic cleavage region is identified; the residues of deleted 1794NANINNANN are shown. The PEST sequence, 1762DTESPS, is also shown. Solid red circles represent some of the putative PKA phosphorylation sites. (B) Putative PKA phosphorylation sites in rabbit α1C. Residues in red are predicted phosphorylation sites and were mutated to Ala in the 17-mutant or 22-mutant transgenic mice. Ser and Thr, shown in blue, although not predicted to be phosphorylated residues, were mutated to Ala. Conserved indicates conserved in the human, guinea pig, rabbit, rat, and mouse. Predicted indicates predicted by at least one of the prediction tools, protein kinase A phosphorylation sites using the simplified kinase binding model (pKaPS), Disorder-Enhanced Phosphorylation Sites Predictor (DISPHOS), GPS, NetPhos, and Scansite. The borderline indicates within the “twilight” zone of pKaPS (31). (C) Schematic representation of the binary transgene system. The αMHC-rtTA is the standard cardiac-specific reverse tetracycline-controlled transactivator system. The αMHCMOD construct is a modified αMHC promoter containing the tet operon (tetO) for regulated expression of FLAG-tagged DHP-resistant (DHP*) α1C.