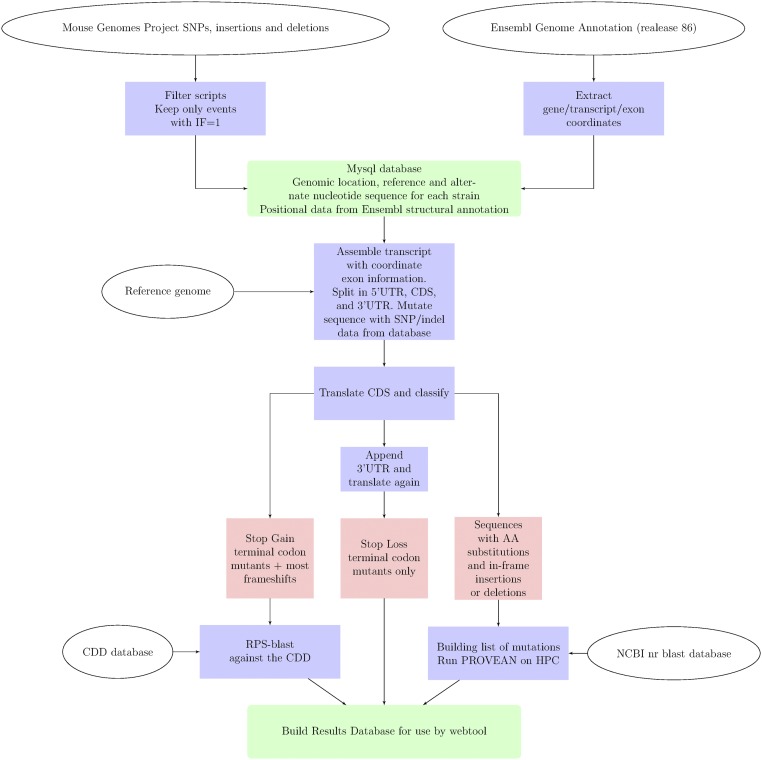
Fig. 1.
Schematic representation of the workflow used to process the sequences of the MGP of the Wellcome Trust Sanger Institute variation data to the final results, which can be queried by the web tool on mousepost.be. The MGP data were processed to only retain high-quality sequence hits, as described in Materials and Methods. Only the genomic location, reference sequence, and sequence in all other strains were kept. In parallel, the Ensembl Genome Annotation for mouse was processed into a table with the genomic coordinates of genes, transcripts and exons. Both the processed MGP and Ensembl tables were stored in a mysql database and indexed for fast access. The annotation tool uses this database in conjunction with the mm10 reference genome to construct the reference 5′ UTR, CDS, and 3′ UTR sequences and then change them to the actual sequences present in the strain of interest. CDS sequences are then translated to protein and compared with the reference sequence for classification. In the case of a SL, the 3′ UTR sequence is added to the canonical CDS to detect the new stop codon, if any. Stop gain genes are checked for lost protein domains by performing an RPS-blast against the conserved domain database from NCBI (www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Mutated sequences have all changed positions scored by PROVEAN to predict the effect of the amino acid substitutions, insertions, or deletions. PROVEAN was run on a computer cluster using a local version of the NCBI database of nonredundant protein sequences. Finally, all results were placed in a mysql database (one table per class), which serves as the back-end for the web tool on mousepost.be.