Figure 2. Transcription-permissive H3K9me2 helps recruit RNAi and precedes H3K9me3 establishment.
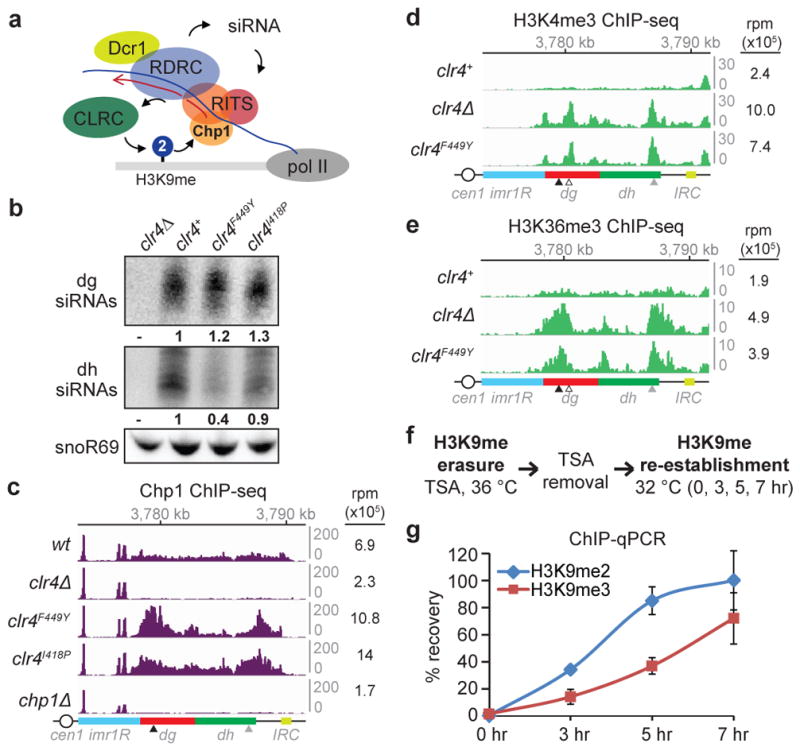
a, Model for recruitment of the RNAi machinery (RITS, RDRC, and Dcr1) and Clr4-containing CLRC to a nascent pericentromeric transcript. b, Northern blot of dg and dh siRNAs in cells with the indicated genotypes. Ratios are determined by fold increase in siRNA levels in mutants over wild-type (wt) cells. snoR69 was used as an internal control. For gel source data, see Supplementary Figure 1. Image represents 3 (dg siRNA) or 2 (dh siRNA) individual experiments. c, Chp1 ChIP-seq reads mapped to the pericentromeric repeat regions on the right arm of chromosome 1. Right, sum of normalized reads mapped to pericentromeric region to the right of cen1. Data is presented as reads per million (rpm, Y axis). Arrows indicate primer locations for ChIP-qPCR analysis in Extended Data Fig. 5. d, e, Same as c, but showing H3K4me3 (d) and H3K36me3 (e) ChIP-seq reads. Empty arrow indicates location for dg2 primers in Extended Data Fig. 5. f, Experimental strategy for de novo H3K9me establishment. g, ChIP-qPCR data showing the recovery kinetics of H3K9me2 (blue) and H3K9me3 (red) at the dh2 pericentromeric repeat. For each time point, H3K9me levels were normalized to that of untreated cells. Error bars, s.d.; n = 3 biological replicates.
