Figure 4. H3K9me3 is required for epigenetic inheritance.
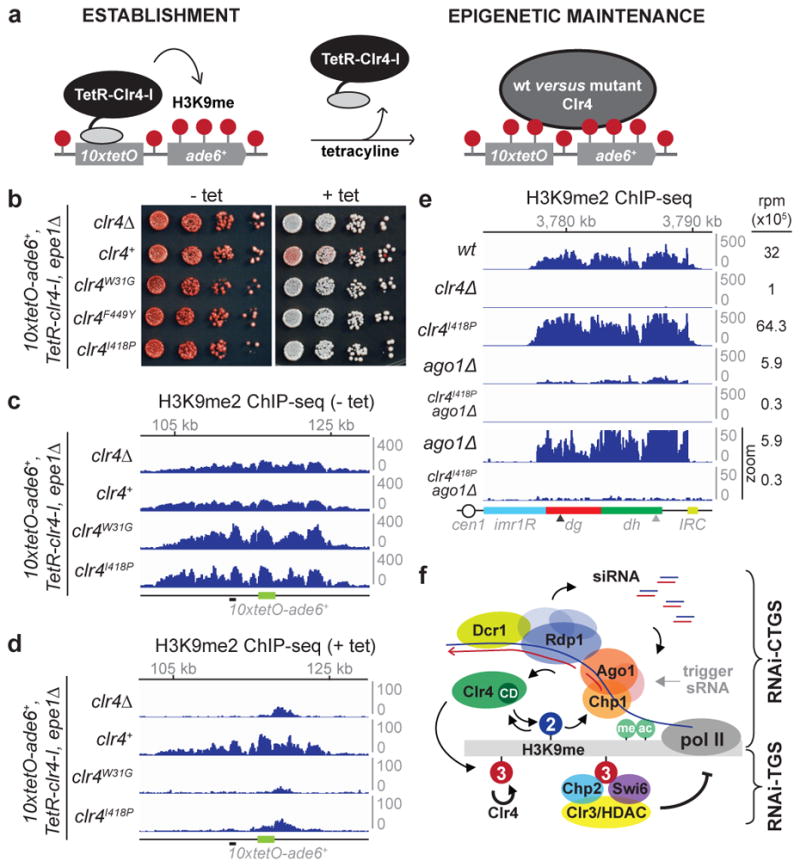
a, Experimental strategy for testing requirements for epigenetic inheritance. b, Silencing assays of 10xtetO-ade6+ on low-adenine medium lacking tetracycline (− tet) or containing tetracycline (+ tet) to assess establishment and maintenance, respectively, in epe1Δ cells, which either lack endogenous clr4+ (clr4Δ), or contain clr4+, clr4W31G, or clr4I418P alleles. Image represents 3 individual experiments. c, H3K9me2 ChIP-seq reads mapped to the 10xtetO-ade6+ region. Both 10xtetO-ade6+ (green) and mug135+ (black dash) located 5 kb upstream of 10xtetO-ade6+ were used for ChIP-qPCR analysis (see Extended Data Fig. 10). d, Same as c, but after 24 hours of growth in + tet medium. e, H3K9me2 ChIP-seq reads mapped to pericentromeric repeats on the right of chromosome 1. The sum of normalized reads is indicated on the right. Data is presented as reads per million (rpm, Y axis). The bottom two tracks have a 10-fold expanded Y axis scale to highlight the complete loss of H3K9me2 in the clr4I418P ago1Δ double mutant cells. f, Schematic summary of the unique roles of H3K9 methylation states. Top: H3K9me2 mediates co-transcriptional degradation of nascent transcripts (RNAi-CTGS) and H3K9me spreading. Bottom: The formation of H3K9me3 domains, which requires the chromo domain (CD) of Clr4, results in efficient recruitment of HP1 proteins and transcriptional gene silencing (RNAi-TGS). H3K9me3, but not H3K9me2, can be epigenetically inherited. See Extended Data section for additional discussion.
