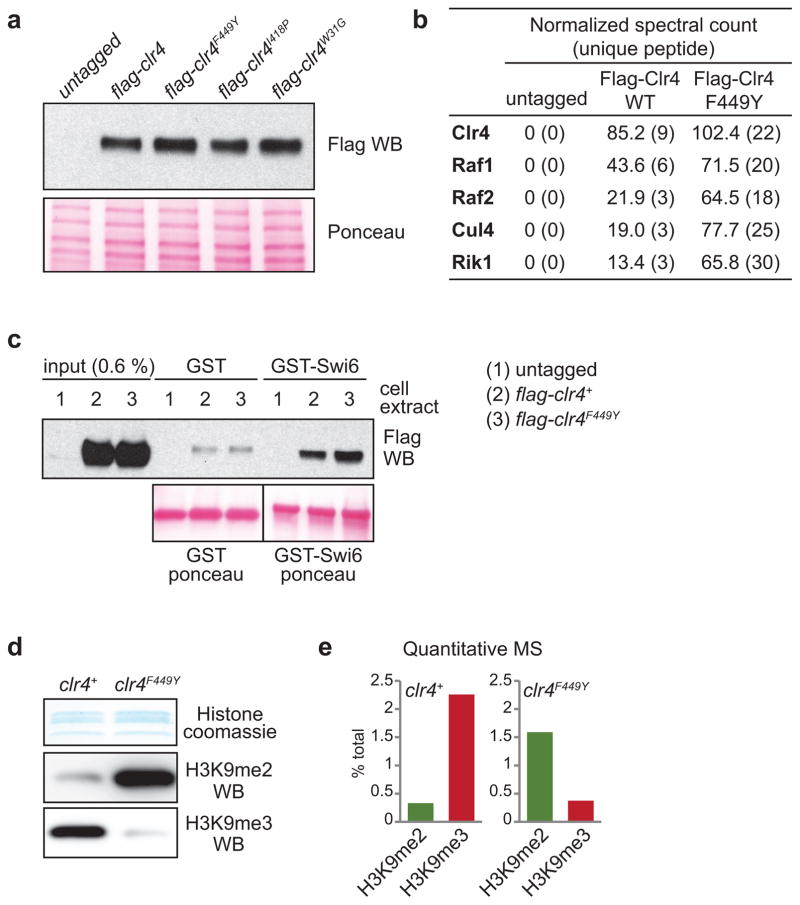
Extended Data Figure 1. Analysis of mutant Clr4 proteins and their effect on histone H3K9me.
a, Western blot of N-terminal 3xFlag-tagged Clr4 showing that SET mutations (F449Y or I418P) or a chromo domain mutation (W31G) do not affect Clr4 protein stability (top). The same blot stained with Ponceau dye is shown as a loading control (bottom). Image represents 2 individual experiments. b, Flag purification of wild-type Clr4 and Clr4F449Y showing both proteins are incorporated into the CLRC methyltransferase complex. c, Pull-down assays showing that wild-type Clr4 and Clr4F449Y interact with recombinant GST-Swi6 with similar efficiency. d, Coomassie staining (top) and western blot (bottom) of histones enriched for H3K9me using Swi6 affinity pull-down from wild-type and clr4F449Y cells showing that Clr4F449Y primarily catalyzes H3K9me2. Image represents 3 individual experiments. e, Quantitative mass spectrometry of histones showing the redistribution of H3K9 methylation states in clr4+ and clr4F449Y cells. The histones were isolated using Swi6 affinity pull-down to increase detection sensitivity. See Extended Data Fig. 6 for quantitative mass spectrometry of H3 tail modifications in total wild-type histones. For gel source data, see Supplementary Figure 1.