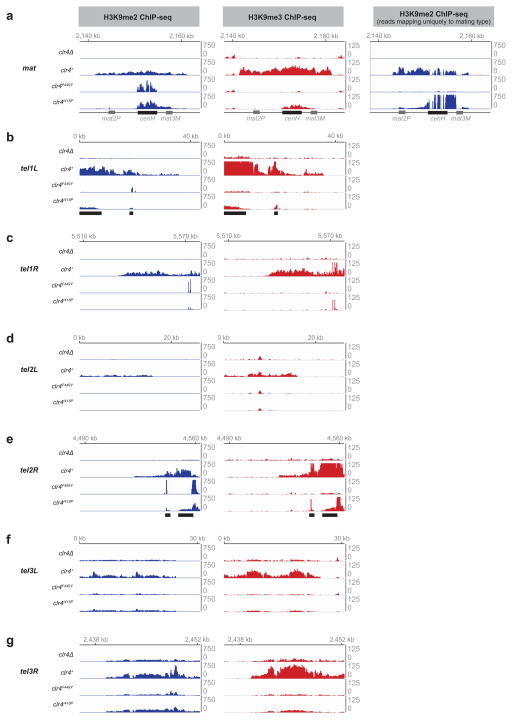
Extended Data Figure 3. Clr4 mutants have reduced H3K9me spreading at mating type and telomeric regions.
a–g, ChIP-seq data showing changes in H3K9me2 and H3K9me3 levels outside of RNAi-dependent nucleation regions (indicated by solid black bars below tracks) at mating type (mat) and telomeric DNA regions (tel1L, tel1R, tel2L, tel2R, tel3L, and tel3R) in clr4Δ, clr4+, clr4F449Y, and clr4I418P cells. tel3L and tel3R represent reads from the rDNA repeats. H3K9me2 reads were randomly assigned to repeated sequences. The reads at cenH are therefore shared with those at the pericentromeric dg and dh repeats (with which cenH shares 98% sequence identity); H3K9me2 reads that map uniquely to the mating type locus are shown (a, right panel). When only unique reads are mapped, a large fraction of total reads corresponding to repeated sequences at centromeres, telomeres, and the mat locus, are removed. This changes the normalized peak heights, which are affected by fewer total mapped reads. Data is presented as reads per million (Y axis).