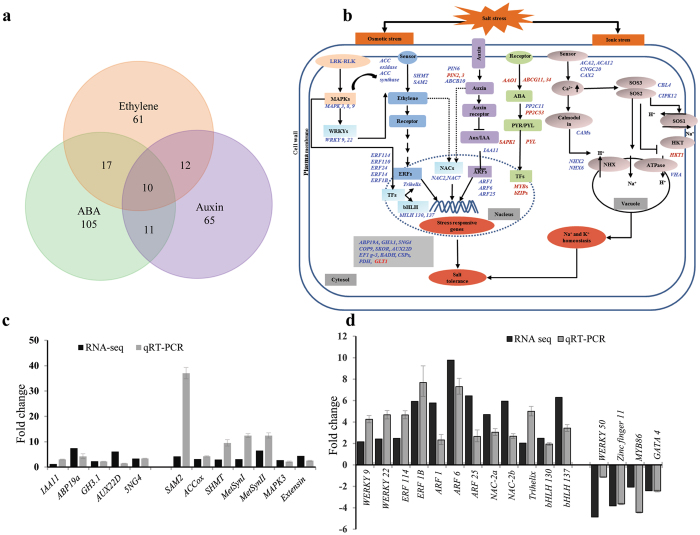
Figure 2.
Phytohormone (ethylene and auxin) signaling-related genes are up-regulated upon salt treatment in A. officinalis. (a) Venn diagram represents the number ABA-, ethylene- and auxin-responsive DEGs identified in A. officinalis transcriptome analysis. (b) Signaling pathways mediating salt tolerance in A. officinalis roots: Major phytohormone (auxin, ethylene and ABA) and Ca2+ signaling pathways that are operative in various plants to render salt tolerance are depicted in the picture. Genes that are up-regulated in A. officinalis roots are indicated in blue, while the down-regulated genes are indicated in red. (c) Expression pattern of some of the genes related to ethylene- and auxin-signaling and (d) expression analysis of DEGs predicted to be encoding transcription factors. Black bar indicates transcript abundance changes calculated by RPKM method. The grey bars plotted with error bars represent the relative expression levels quantified by qRT-PCR method. Relative expression levels of transcripts with reference to Ubiquitin 10 transcript levels are plotted, qRT-PCR data represent means ± SD, from 3 biological replicates. IAA11: Auxin-responsive protein11, ABP19a: Auxin-binding protein, GH3.1: Probable indole-3-acetic acid-amido synthetase, AUX22D: Auxin-induced protein 22D, 5NG4: Auxin-induced protein 5NG4, SAM2: S-adenosylmethionine synthase 2, ACCox: 1-aminocyclopropane-1-carboxylate oxidase homolog 1, SHMT: Serine hydroxymethyltransferase, Metsyn: 5-methyltetrahydropteroyltriglutamate–homocysteine methyltransferase, MAPK3: Mitogen-activated protein kinase3.