Fig. 3.
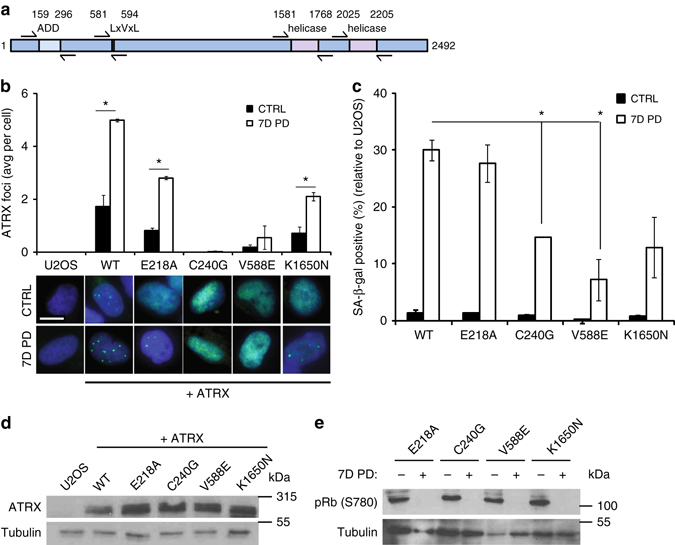
The ability of ATRX to support CDK4i-induced senescence in U2OS cells requires HP1 and H3K9me3 binding sites. a A schematic of ATRX indicating the domains and amino acid residues numbered as annotated on UniProt is shown. Arrows indicate the paired sequencing primers used to confirm the mutations. b–d U2OS cells were transfected with vectors expressing wild type or mutant ATRX alleles and stable transformants were selected with G418 and sorted to recover a GFP-low population as described in the methods. WT indicates wild type. b ATRX immunofluorescence was carried out in the mutants described in the legend to Fig. 2. The average number of ATRX foci per cell is plotted and representative images are shown. c The accumulation of SA-β-gal-positive cells was scored 7 days after PD0332991 (PD) treatment. Asterisk indicates p < 0.05 for the induction of SA-β-gal following treatment compared to cells expressing wild type ATRX. p-values were calculated using a two-sided Student’s t-test. d ATRX was detected with the A301-045A antibody. Tubulin is a loading control. e Rb phosphorylation, a measure of cell proliferation, was measured in the U2OS cells expressing different mutant ATRX constructs before and after treatment with PD0332991 (PD) for 7 days. Tubulin is a loading control. All data are represented as mean ± SEM from at least two independent experiments. See also Supplementary Fig. 10 for uncropped blots. All images were taken at the same magnification. The scale bar is 20 microns
