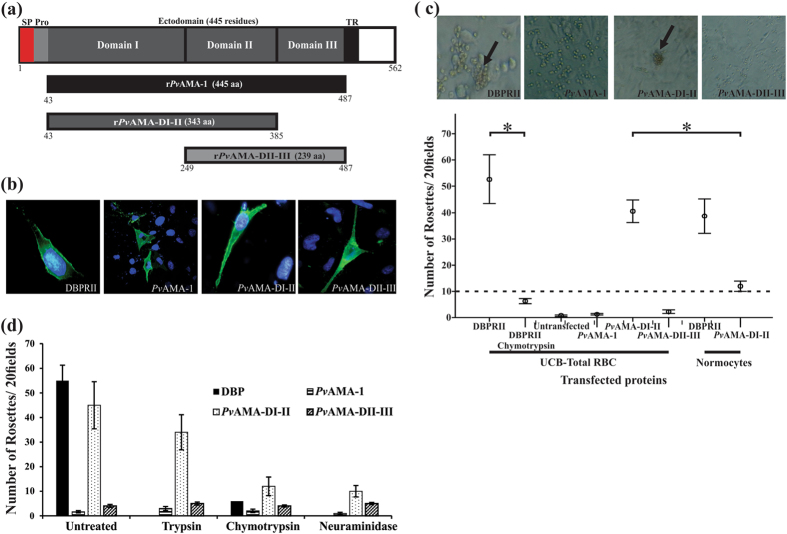
Figure 1.
COS7 cells expressing PvAMA-DI-DII bound to UCB RBC. (a) Schematic representation of PvAMA-1 primary structure. Signal peptide (SP) is indicated in red, pro-sequence (Pro) in light grey, ectodomain (consisting of domain I (DI), domain II (DII) and domain III (DIII)) in dark grey, transmembrane region (TR) in black and cytoplasmic tail in white. The three fragments were expressed both on COS-7 cell membrane and as soluble recombinant proteins in E. coli. Black shows the complete PvAMA-1 ectodomain (rPvAMA-1), dark grey represents domains I-II (rPvAMA-DI-II) and light grey domains II–III (rPvAMA-DII-III). The first and last residues in each region are indicated. (b) Immunofluorescence assay. Each of the three PvAMA-1 fragments was cloned in the pRE4 vector and expressed on COS-7 cell membrane. The presence of HSV gD protein signal peptide and transmembrane domain in pRE4 vector enabled the expression of each fragment on cells surface, as shown by green fluorescence. Cell nuclei were stained with DAPI. (c) Rosette formation assays. The binding assay was performed with UCB RBC or reticulocyte-depleted normocytes. Only PvAMA-DI-DII and DBPRII transfected cells could form rosettes (black arrows).The amount of rosettes for each protein expressed on COS-7 cells is shown. DBPRII was used as positive control while non-transfected cells and DBPRII binding to chymotrypsin-treated UCB were used as negative controls. (d) Trypsin-, chymotrypsin- and neuraminidase-treated UCB RBC binding assays. It can be seen that the amount of rosettes became reduced with PvDI-DII protein when RBC were treated with chymotrypsin and neuraminidase. Only intact erythrocyte and chymotrypsin-treated DBPRII binding was evaluated. At least three independent experiments (with replicates) were performed for all experiments. The error bars indicate the standard deviation.