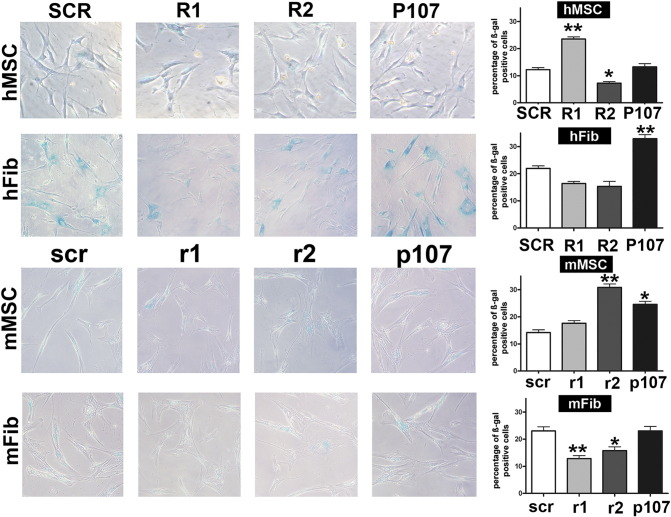
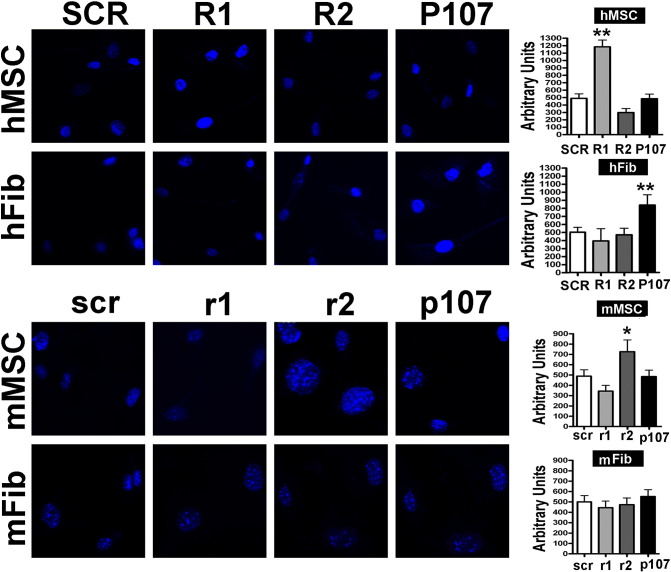
Figure 2.
(A) Senescence levels in cells with silenced retinoblastoma proteins.
The presence of senescent cells was evaluated in human and mouse cells following the silencing of RB1, RB2/P130, and P107 with specific shRNAs. The picture shows representative microscopic fields of senescence-associated beta-galactosidase–positive cells in the different experimental conditions. The histograms show the percentage of senescent cells in MSCs and fibroblasts from both human and mouse origin (hMSC, mMSC, hFib, and mFib, respectively). R1, R2m and P107 stand for shRNAs against human RB1, RB2/P130, and P107 mRNAs, respectively. The control shRNA with a scrambled sequence was named SCR. The shRNAs against the corresponding mouse mRNAs were indicated as r1, r2, and p107, respectively. Control shRNAs for mouse cells was named scr. Data are expressed with standard deviation (n = 3, *P < .05, **P < .01).
(B) DAPI staining.
Fluorescence photomicrographs show cells stained with DAPI (blue). Representative microscopic fields are shown. The graph shows the degree of DAPI staining. For each positive cell, the DAPI intensity was acquired with a CCD camera and analyzed with Quantity One 1-D analysis software (Bio-Rad Laboratories). We calculated the sum of the fluorescent pixel values of DAPI-positive cells and then determined the average fluorescent pixel intensity, which was expressed in arbitrary units. For every experimental condition, staining intensity was determined for 200 cells. Data are expressed with standard deviation (n = 3, *P < .05, **P < .01).