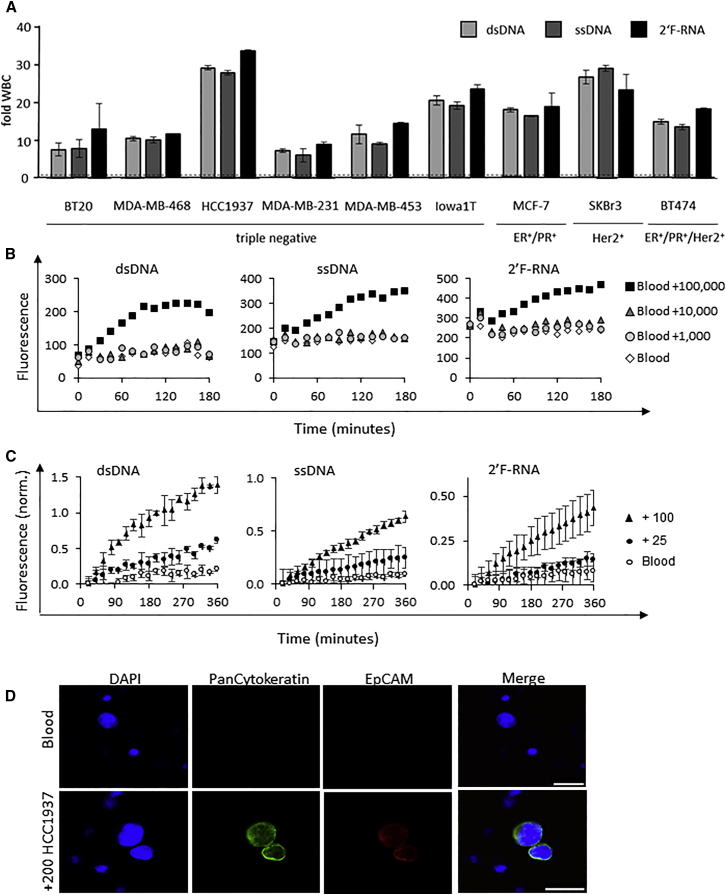
Figure 4.
Nuclease-Activated Probe Assay Specificity
(A) Nuclease-activated probes (dsDNA, ssDNA, and 2′F-RNA) were incubated with lysates of 100 white blood cells isolated from the blood of a healthy donor or various BCa cell lines (BT20, MDA-MB-468, HCC1937, MDA-MB-231, MDA-MB-453, Iowa1T, MCF7, SKBr-3, and BT474). Fold WBC is defined as the fluorescence signal generated from each individual BCa cell line divided by that generated by the white blood cells. Fluorescence was measured in a microplate reader every 20 min for a total of 6 hr. The 4-hr time point is plotted. (B) Sensitivity of detection of cancer cells in blood without prior cancer cell capture and/or enrichment step. A defined amount of healthy donor blood (about 1 mL) with the addition of increasing amounts of MDA-MB-453 cells (1–100,000 cells per 1 mL of blood) was subjected to the nuclease-activated probe assay using the three nuclease-activated probes (dsDNA, ssDNA, and 2′F-RNA). (C) Nuclease-activated probe assay sensitivity. Blood samples from a healthy donor were spiked with either 100 or 25 HCC1937 cancer cells per milliliter of blood, and samples were evaluated using the nuclease-activated probe assay. Blood, not spiked with cancer cells, served as a control (blood). The signal intensity was normalized to the background fluorescence of the corresponding probe. (D) Blood samples from a healthy donor with and without the addition of 200 HCC1937 cancer cells per milliliter of blood. Cancer cells in blood were immunostained with mouse anti-human PanCytokeratin (green) and rat anti-EpCAM (red). Scale bars: 25 μm.