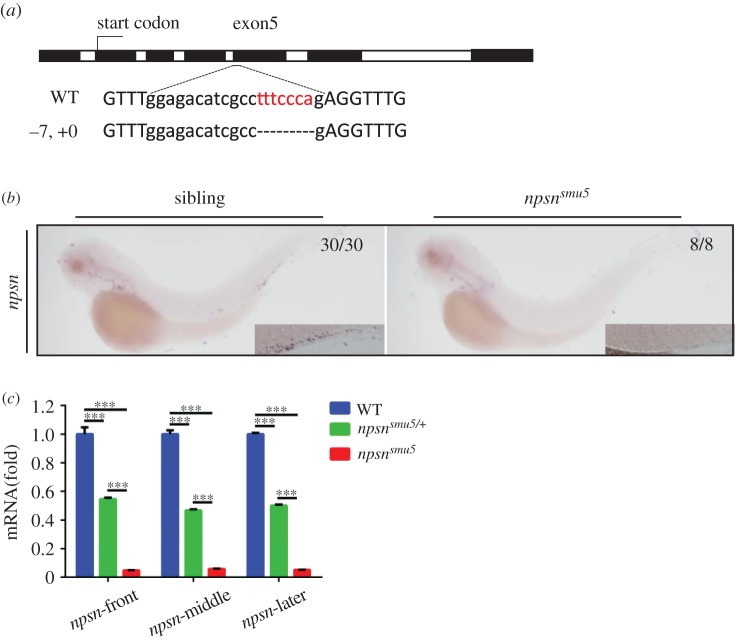
Figure 2.
Generation and identification of nps-deficient zebrafish. (a) The CRISPR/Cas9 system was used to generate npsn-knockout zebrafish. The Cas9 target was chosen as exon5 of npsn. Mutants were selected using T7E1 enzyme digestion and confirmed using Sanger sequencing. In F1 founders, a mutant line (−7, +0) containing a frameshift mutation was obtained and retained for follow-up studies. (b and c) Degradation of npsn mRNA in npsnsmu5 mutant embryos. (b) WISH results for npsn mRNA indicated significant decreases in npsnsmu5 mutant embryos at 3 dpf. (c) qRT-PCR results indicating that npsn mRNA expression was downregulated to 5% in npsnsmu5 mutant embryos. (Mean ± s.e.m., n = 20 per experiment, triplicated). Statistical significance was determined by one-way ANOVA. ***p < 0.001.