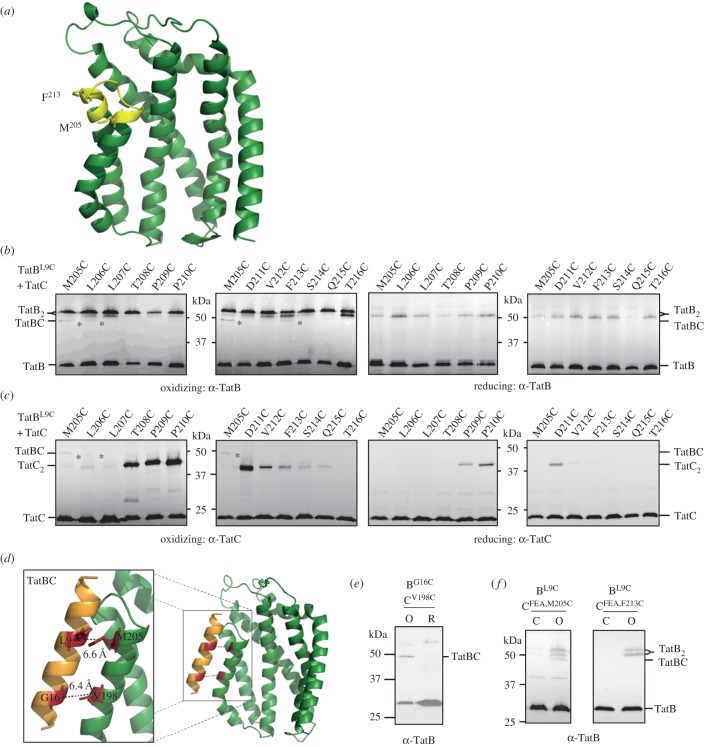
Figure 2.
TatBL9C interacts with TatCM205C in vivo. (a) Homology model of E. coli TatC showing positions of the residues used for disulfide cross-linking analysis in yellow. The side-chains of M205 and F213 are indicated. (b,c) Western blot analysis (separated on 10% polyacrylamide gels) of membranes from E. coli strain MC4100ΔBC producing TatBL9C alongside the indicated Cys substitutions in TatC (from plasmid p101C*BC) following exposure of whole cells to 1.8 mM CuP (oxidizing) or 10 mM DTT (reducing) for 1 min. Cross-linked products were visualized by immunoblotting using (b) an anti-TatB peptide antibody or (c) an anti-TatC antibody. The asterisks indicate likely TatBC cross-links. (d) Structural model of TatB interacting with TatC at the polar cluster site (adapted from [15]). The backbone distances between TatBL9/TatCM205 and TatBG16/TatCV198 are shown. (e) Whole cells of strain MC4100ΔBC producing TatBL9C alongside TatCF94A,E103,/M205C or TatCF94A/E103A/F213C from plasmid p101C*BC (annotated TatCFEA,M205C or TatCFEA,F213C, respectively) were left untreated (C) or incubated for 1 min with 1.8 mM CuP (O) as indicated. Following membrane preparation, cross-links were detected with an anti-TatB peptide antibody.