Fig. 2.
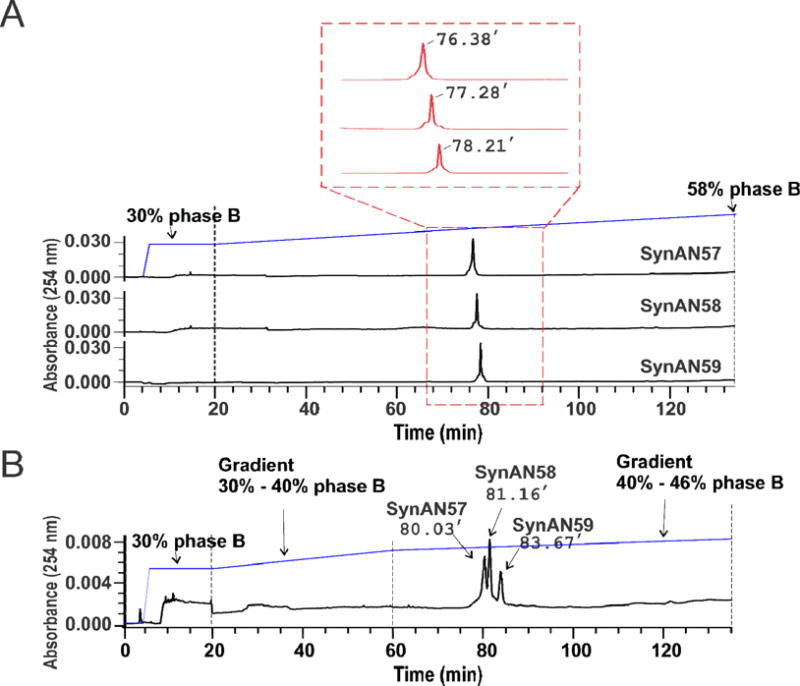
Single-nucleotide separation of RNAs up to 59 nt by HPLC. (A) Individual chromatogram for SynAN57, SynAN58 and SynAN59. Each sample was run through an XBridge C18 reverse-phase column at 50 °C (see detail in Methods). The middle section framed by the red box shows a detailed peak profile and retention times. The running buffer contained 16.3 mM of TEA and 400 mM of HFIP at pH 7.9 with 5% methanol (Phase A) or 30% methanol (Phase B) (see Methods). The gradient started at 30% of Phase B and ended at 58% of Phase B in 115 minutes. Unless otherwise noted, the flow rate of all experiments was kept at 0.5 ml/minute and the temperature of column was maintained at 50 °C. (B) An equal molar SynAN57, SynAN58, and SynAN59 were mixed and run through the same XBridge column. The retention times of three RNAs, from 57-nt to 59-nt, are marked. For elution, two sections of linear gradient were used to improve the separation. The first linear gradient started at 30% of Phase B and ended at 40% in 40 minutes, and the second linear gradient began at 40% of Phase B and ended at 46% of Phase B in 75 minutes. The flow rate was kept at 0.5 ml/minute.
