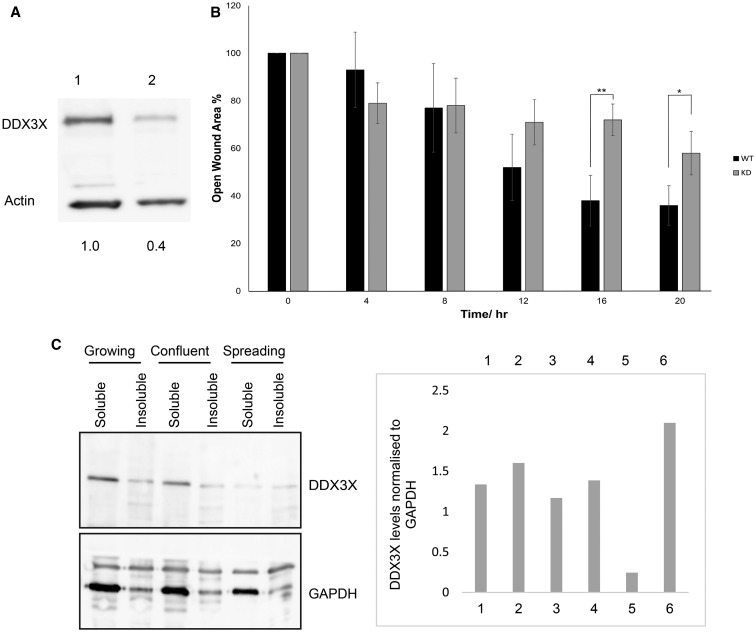
Figure 1. Reduction in DDX3X levels impedes cell migration.
(A) Aliquots of extract (20 µg of total protein) from parental MRC5 (lane 1) or MRC5 cells partially depleted of DDX3X by CRISPR/Cas9 (lane 2) were resolved by SDS–PAGE and visualised using immunoblotting. Levels of DDX3X expression were quantified relative to actin (set at 1.0). (B) Wound-healing assays were carried out as described using parental (WT) and DDX3X-depleted (KD) cells over a period of 20 h. Following removal of the bung from confluent cultures, wound closure was measured using the Image J software and expressed as % of open wound area relative to time 0. Data are presented as the means ± SD, n = 3. All data were analysed using a one-way ANOVA test; *P ≤ 0.05; **P ≤ 0.01. (C) MRC5 cells were grown to either logarithmic or stationary growth phases, washed in PBS and incubated with digitonin to extract soluble protein, as described in the Materials and Methods section. For spreading cells, cells were harvested during logarithmic growth using trypsin and allowed to re-settle and spread on dishes until lamellipodia were established. Cells were scraped into lysis buffer and lysed to extract soluble and non-soluble protein. Upper panel: aliquots of protein (5 µg of total protein, ∼1/300 of the total soluble fraction and 1/50 of the insoluble fraction) were separated by SDS–PAGE and proteins were visualised by Western blotting. DDX3X levels were normalised to GAPDH (lower panel).