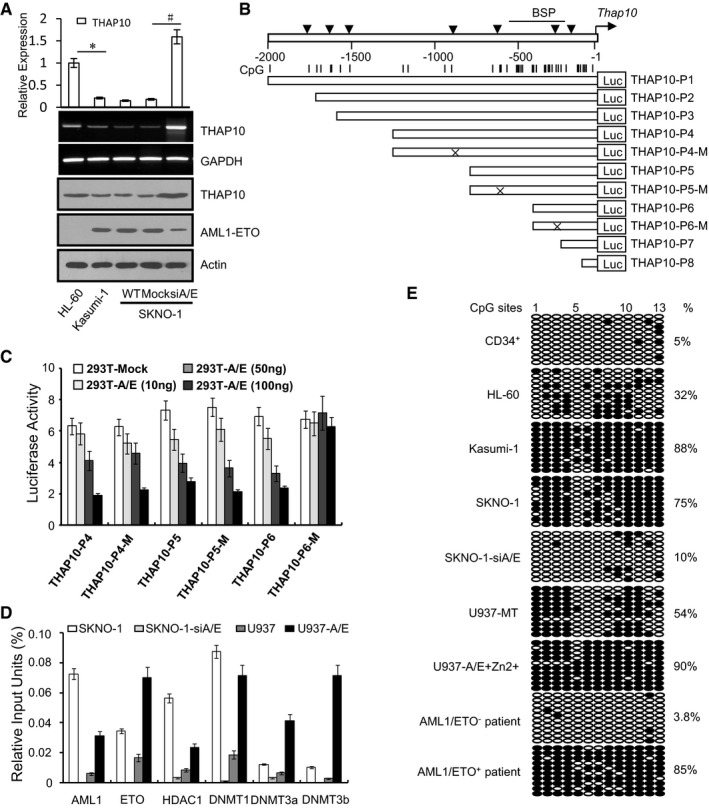
Upper panel: Relative quantification of THAP10 mRNA levels in the indicated leukaemia cell lines. The results represent the mean of three independent evaluations ± SD (*P = 0.001, #
P = 0.0007, Student's t‐test was used for the comparisons). Middle and lower panels: mRNA and protein levels of THAP10 and AML1‐ETO, respectively.
Upper panel: Schematic diagrams of the AML1‐binding sites and the CpG islands along the THAP10 genes. Numbers indicate the nucleotides relative to pre‐THAP10 (−1 nt). Vertical arrowheads indicate the AML1‐binding sites; vertical lines indicate CpG dinucleotides; horizontal bar illustrates the regions analysed by bisulphite sequencing. Lower panels: A series of constructs containing different AML1‐binding sites and their mutants.
Luciferase reporter activities of human 293T cells transiently co‐transfected for 48 h with luciferase reporters containing the wild‐type sequence of the THAP10 promoter or its mutant counterparts, together with increasing amounts of pcDNA3.0 vectors containing AML1‐ETO or mock cDNA (293T‐Mock). Triplicate values are defined as mean ± SD.
Chromatin immunoprecipitation (ChIP) using the indicated antibodies or IgG, after which qRT–PCR was performed to evaluate the specificity of protein binding. Triplicate values are defined as mean ± SD.
Genomic bisulphite sequencing to detect the methylation status of the DNA sequences surrounding the AML1‐binding site (−280 nt) in the pre‐THAP10 gene upstream region in CD34+ haematopoietic progenitors isolated from peripheral blood (PB) of healthy donors, the indicated leukaemia cell lines and primary leukaemic blasts. Each row of circles represents the sequence of an individual clone. Solid and empty circles represent methylated and unmethylated CpG dinucleotides, respectively.
Data information: All experiments were performed in triplicate.