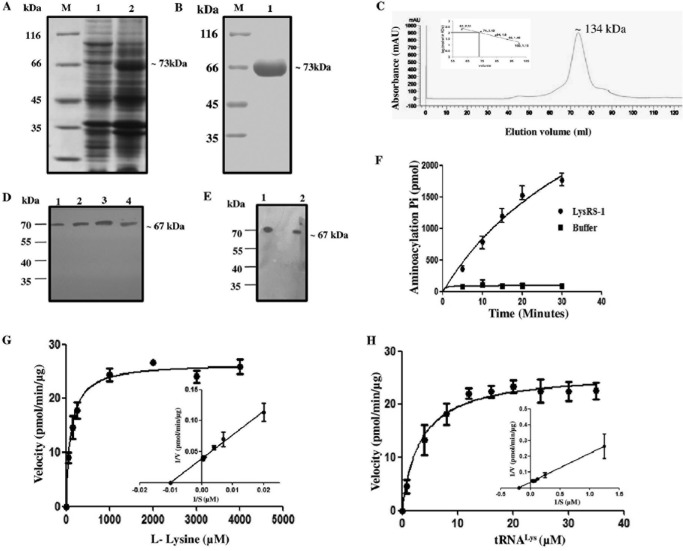
FIG 3 .
Protein induction, purification, and enzymatic characterization of recombinant LdLysRS-1. (A) SDS-PAGE analysis of whole-cell lysate of uninduced and induced E. coli BL21(DE3) cells transformed with pET-30a–LdLysRS-1. M, molecular mass marker; lane 1, uninduced bacterial cell lysate; lane 2, induced bacterial cell lysate. (B) Purification of rLdLysRS-1 protein on Ni2+-NTA affinity resin. M, molecular mass marker; lane 1, eluted fraction with 100 mM imidazole showing purified rLdLysRS-1. (C) GPC elution profile of purified LdLysRS-1. Comparison with standard markers indicates that LdLysRS-1 elutes at a size corresponding to the dimeric state. mAU, milli-absorbance unit. (D) Western blot analysis of the rLdLysRS-1 protein and promastigote cell lysates of wild-type (WT) parasites using anti-LdLysRS-1 antibody. Lane 1, 0.5 μg rLdLysRS-1 protein; lane 2, 1 μg rLdLysRS-1 protein; lane 3, 2 μg rLdLysRS-1 protein; lane 4, Leishmania promastigote cell lysate (~40 μg). (E) Western blot analysis of the rLdLysRS-1 protein and amastigote cell lysates of WT parasites. Lane 1, 2 μg recombinant LdLysRS-1 protein; lane 2, Leishmania amastigote cell lysate (~40 μg). (F) Time course of tRNALys aminoacylation by recombinant LdLysRS-1. Reactions were performed with l-lysine and tRNALys as the substrates. The data show an average from three experiments performed in duplicate ± SD. (G and H) Aminoacylation kinetics of LdLysRS-1 as a function of l-lysine concentration (G) and tRNALys concentration (H). The results represent means ± SD (n = 3).