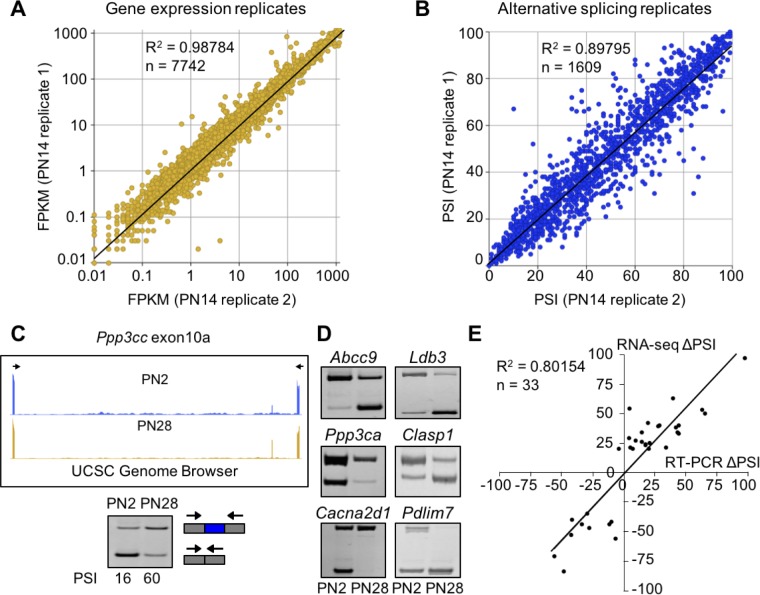
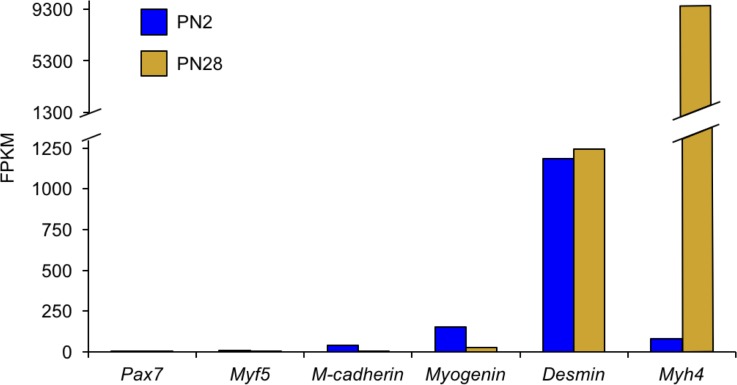
Figure 1. RNA-seq data is high quality and reproducible.
(A and B) Biological replicates for PN14 were compared to analyze variation in gene expression (A) and alternative splicing (B) using Cufflinks and MISO, respectively. (C) RT-PCR to quantitate splicing use primers that anneal to the constitutive exons flanking an alternative exon, displayed on the UCSC Genome Browser (above) to determine the percent spliced in (PSI). (D) RT-PCR splice products for alternative splicing events comparing PN2 to PN28. (E) Plot comparing PN2 to PN28 RNA-seq ΔPSI values and ΔPSI values obtained by RT-PCR.