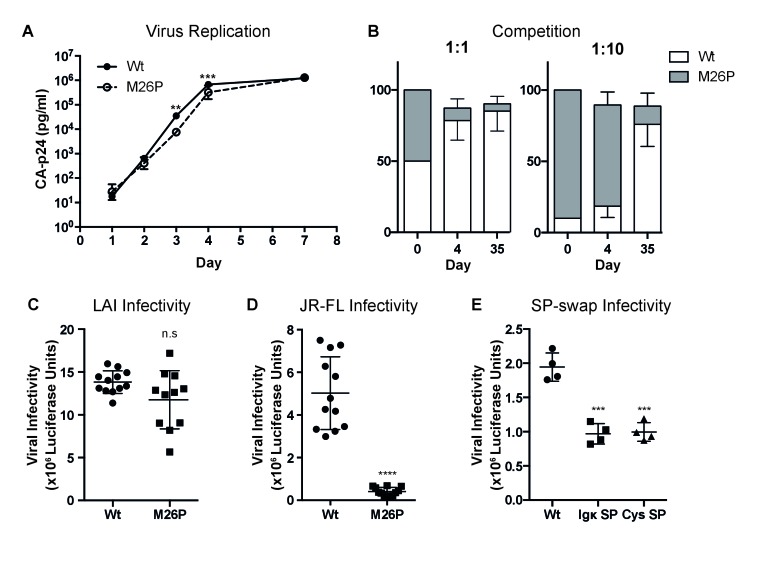
Figure 8. Co-translational signal-peptide cleavage causes functional gp160 defects.
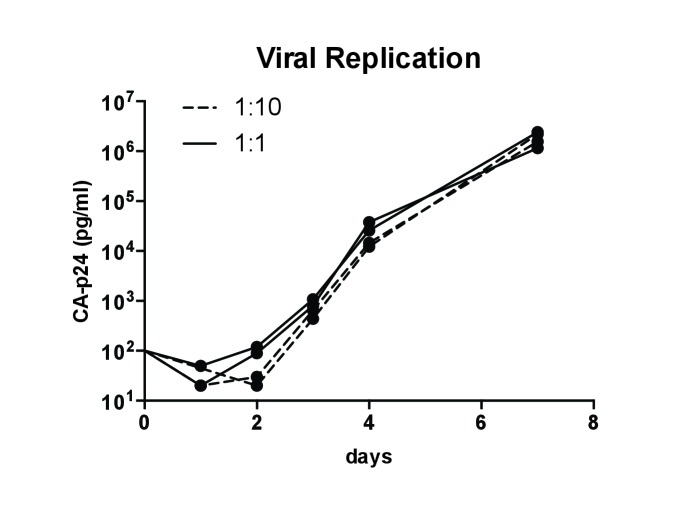
(A) SupT1 cells (5x106) were infected with 20 µg of wild-type or M26P-mutant pLAI DNA constructs using electroporation. Replication was monitored by CA-p24 ELISA. The replication curves shown represent the averaged values of 6 independent experiments. p=0.0058 (**) and 0.003 (***). (B) SupT1 cells (Krogh et al., 2001) were infected with a total of 50 pg CA-p24 equivalent wild-type or M26P-mutant LAI virus (produced by transfected HEK293T cells) in ratios 1:1 and 1:10, each in duplicate (biological replicate). After 4 and 35 days, the isolated virus from each culture was sequenced and the electropherograms were quantified. (C,D,E) TZM-bl reporter cells were infected with: 500 pg of wild-type or M26P LAI virus (produced by HEK293T cells) (n.s, p=0.0833) (C), 1,000 pg of wild-type or M26P-mutant JR-FL pseudo-virus (produced by HEK293T cells) (p<0.0001). One M26P data point was excluded as it lay well below the background. (D), 1,000 pg of wild-type, Ig κ (p=0.0004) or cystatin-SP (p=0.0005) mutant LAI pseudo-virus (produced by C33A cells) (E) and infectivity was measured by Luciferase activity. The data in panels A, C and D are derived from 3 independent experiments, using 3 independently produced (pseudo-)virus stocks (biological replicates), each performed in quadruplicate (technical replicates). The data in panel B are derived from 2 independent experiments, using 2 independently produced virus stocks (biological replicates) and sequenced with two independent primers (technical replicates). The data in panel E are derived from 1 pseudovirus stock (biological replicate), performed in quadruplicate (technical replicates). All significance values were calculated with an unpaired, two-tailed t test with Welch’s correction.