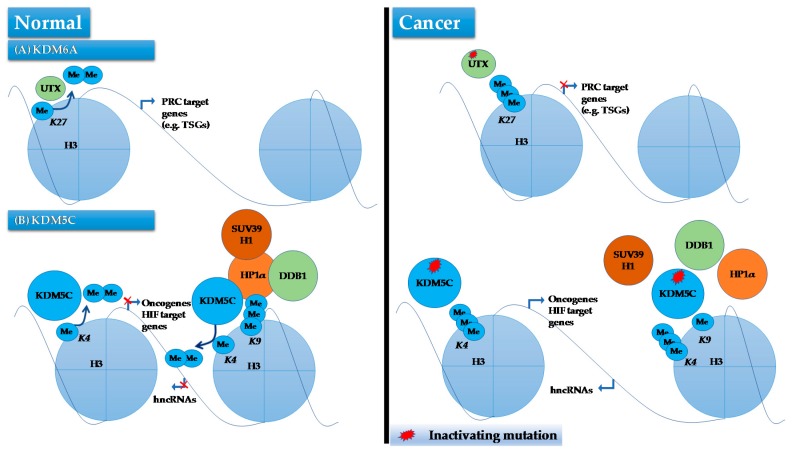
Figure 5.
Functional consequences of KDM6A (or UTX) and KDM5C inactivation. (A) H3K27me3 is a suppressive marks associated with PRC target genes. UTX removes methyl groups from H3K27 and activates PRC target genes including multiple TSGs. Accordingly, loss of UTX in ccRCC contributes to trimethylation of H3K27, leading to inactivation of PRC target genes (e.g., TSGs) [27,144]; (B) KDM5C is a histone demethylase (HDM) that remove methyl groups from H3K4me3. H3K4me3 is a mark of active transcription and the removal of methyl group(s) by KDM5C results in suppression of expression of many genes including HIF target genes. Therefore KDM5C has been suggested to play a role in controlling hypoxia-driven gene expression. In addition, demethylation of H3K4me3 by KDM5C at heterochromatic sites is required for the recruitment of silencing complex consisted of SUV39H1, HP1α, CUL4 and DDB1, which promotes tri-methylation of H3K9 for heterochromatin assembly and silencing of heterochromatic non-coding RNAs (hncRNAs). Inactivation of KDM5C in ccRCC, leads to the overexpression of genes that contribute to tumorigenesis including hypoxia-regulated genes, and uncontrolled expression of hncRNAs, which in turn triggers heterochromatin dysregulation and genome instability [147].