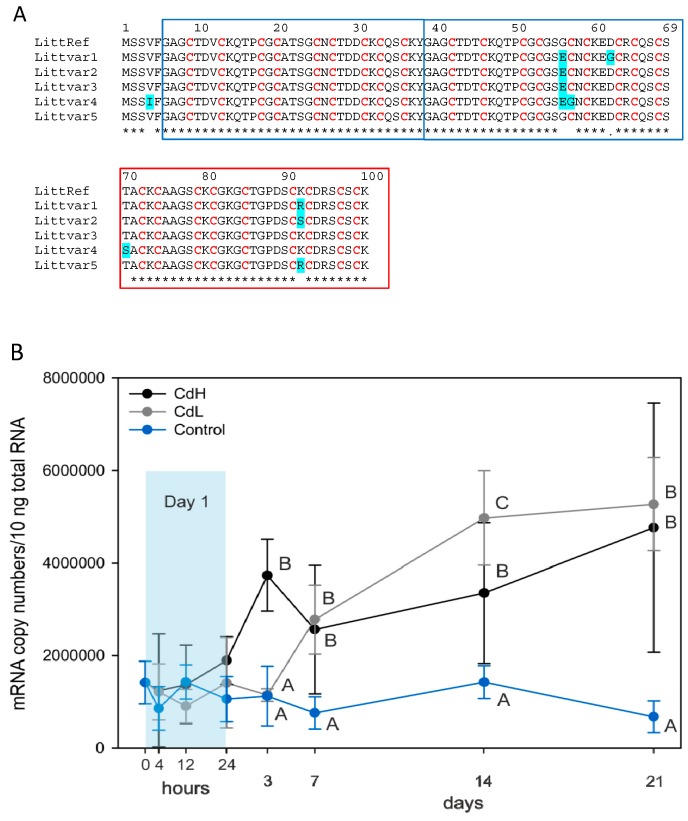
Figure 3.
(A) Translated protein sequences of allelic variants of Littorina littorea, all of them caused by single nucleotide polymorphisms (SNPs) in the coding region of the respective gene. The sequence variants (Littvar1, Littvar2, Littvar3, Littvar4, Littvar5) are aligned with the first reported reference sequence (LittRef) of the Littorina littorea MT (GenBank Acc.Nr.: AY034179.1) [31]. Exchanged amino acid residues in the allelic variants are marked in blue. The tri-partite structure of the protein into different domains is indicated by blue (α domains) and red frames (β domain) [37]. The allelic variant sequences are available from GenBank under the acc. nrs.: KY963497 (Littvar 1), KY963498 (Littvar2), KY963499 (Littvar3), KY963500 (Littvar4) and KY963501 (Littvar5); (B) mRNA transcription of the Cd metallothionein (MT) gene of Littorina littorea in control animals (Control, blue line and symbols) and in winkles exposed to nominal Cd concentrations of 0.25 mg/L (CdL, grey line and symbols) and 1.00 mg/L (CdH, black line and symbols) over a period of 21 days. Mean values and standard deviations (n = 4) are shown. The significance of the mRNA transcription curves was confirmed by ANOVA (p ≤ 0.001). The significant differences of values between Cd-exposed and control individuals at different time points (Holm–Sidak pairwise multiple comparison, significance level 0.05) are indicated by different letters (A, B, C). Sampling points during the first 24 h (Day 1) are under-laid in blue.