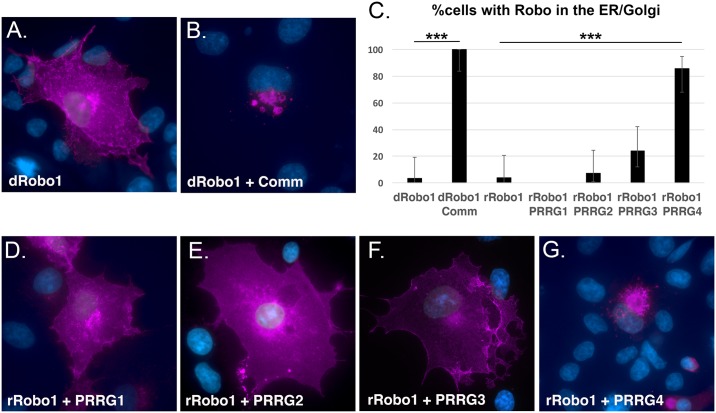
Fig 6. PRRG4 re-localizes rRobo1 from the plasma membrane.
COS cells were transfected with plasmids encoding the genes indicated in the panels and antibody stained with anti-Robo in magenta. Cells were counterstained with DAPI (blue) to reveal the nucleus. A. Drosophila Robo1 (dRobo1) and B. Co-expression of both dRobo1 and Comm leads to re-localization of dRobo1 from the cell surface to the ER/Golgi. C. Quantification of results for re-localization experiments. Cells transfected with the genes indicated were stained for dRobo1 or rRobo1. Subcellular localization at either the plasma membrane or predominantly in the ER/Golgi were scored by an experimenter blind to the plasmids present. At least 22 healthy cells as judged by nuclear staining were scored for each category. The percentage of cells with ER/Golgi localization is shown in the bar graph. Error bars are the 95% confidence interval to reflect sampling noise. Statistical significance relative to dRobo1 and rRobo1 controls is shown (*** p < 0.01, highly statistically significant) and was calculated using the Fisher exact test with two tails. For the PRRG and rRobo1, the Bonferroni correction was applied. Comparison of dRobo1 with and without Comm has a p value < 0.0001. Comparison of rRobo1 and PRRG4 has p < 0.0001. The PRRG3 and rRobo1 data are trending towards statistical significance, p = 0.0538 (cutoff value is p < 0.0125). D. rRobo1 in the presence of PRRG1 is localized predominantly to the cell surface. E. Co-expression of PRRG2 and rRobo1 results in cell surface localization of rRobo1. F. PRRG3 expression can result in a reduction in the level of rRobo1 on the cell surface. G. The majority of cells co-expressing PRRG4 and rRobo1 display an ER/Golgi localization for rRobo1. The underlying data are shown in S2 Data.