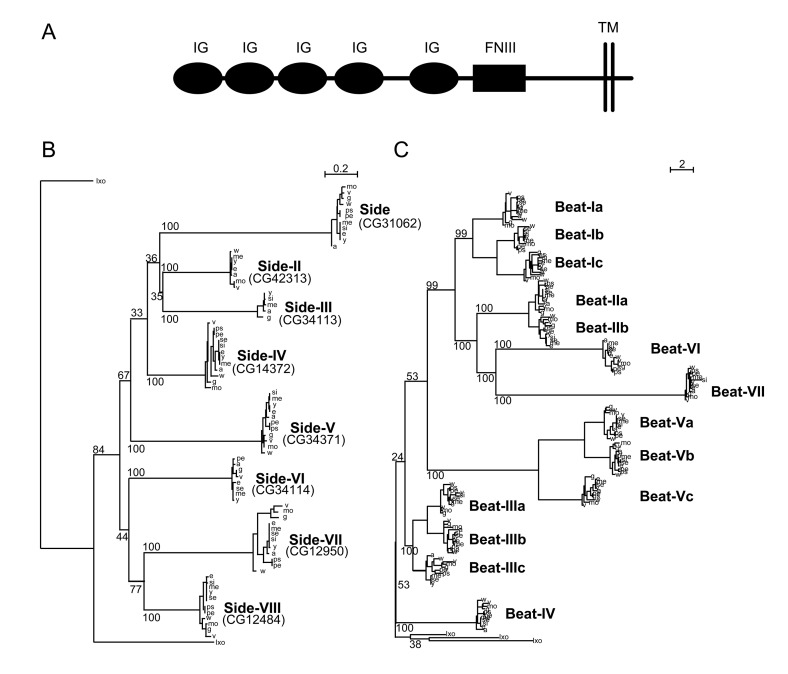
Figure 1. Phylogenetic analysis of Beaten Path and Sidestep paralogs.
(A) Phylogeny of the Beat family of receptors rooted against the tick Ixoides scapularis (Ixo) Beats. Beat-VII and Beat-VI share a more recent ancestor than previously described. (B) Extracellular architecture of the Side subfamily. Detailed ClustalW alignment of individual domains and conservation are in Figure 1—figure supplement 1. (IG, immunoglobulin superfamily; FNIII, Fibronectin type III; TM, transmembrane domain). (C) Phylogeny of the Side family of related proteins rooted against similar IgSF proteins predicted in the tick, Ixoides scapularis (Ixo) that form a distinct outgroup. Names are assigned to the paralogs on the basis of their evolutionary distance from Sidestep and their CG Flybase identifiers are in parentheses.