Abstract
Top-down proteomics is the analysis of intact proteins in their endogenous form without proteolysis, preserving valuable information about post-translation modifications, isoforms and proteolytic processing. The quality of top-down LC-MS/MS datasets is rapidly increasing due to advances in instrumentation and sample processing protocols. However, the top-down mass spectra are substantially more complex compared to conventional bottom-up data. To take full advantage of the increasing data quality, there is an urgent need to develop algorithms and software tools for confident proteoform identification and quantification. In this study, we present a new open source software suite for top-down proteomics analysis consisting of an LC-MS feature finding algorithm, a database search algorithm, and an interactive results viewer. The presented tool along with several other popular tools were evaluated using human-in-mouse xenograft luminal and basal breast tumor samples that are known to have significant differences in protein abundance based on bottom-up analysis.
Introduction
While mass spectrometry (MS) based proteomics has been successful for identifying and quantifying peptides and post-translational modifications (PTMs), the characterization of intact protein forms (i.e. proteoforms) remains challenging1–4. At present, the majority of proteomics studies are peptide-based because intact protein proteomics (or top-down) is more challenging at almost every stage of the analytical process: sample preparation, liquid chromatography (LC) separation, fragmentation, and data analysis5,6. In particular, the challenges and lack of confidence in data analysis are a major reason preventing proteomics researchers from adopting top-down studies7. Unlike traditional bottom-up proteomics, where numerous software tools are available, only a handful of tools are available for top-down characterization, and the data analysis often requires laborious manual interpretation. In this paper, we present a new open source software suite for top-down data analysis, named Informed-Proteomics.
In general, the top-down data analysis workflow consists of three steps: feature deconvolution, protein characterization via database search of fragmentation data, and validation. In every step, there are challenges that make top-down data analysis substantially more difficult than bottom-up data analysis. First, the size of intact proteins means that they typically have higher and more diverse charge states after electrospray ionization. This distributes the ion signal over a broader number of charge states with increasingly large isotopomer envelopes, substantially reducing the signal-to-noise ratio and challenging MS resolution. Detecting ion signals and accurately calculating precursor mass is essential to proteoform identification and quantification. Existing deconvolution tools such as THRASH8, Xtract9 and MS-Deconv10,11 adopt spectrum-centric approaches and create a simplified spectrum of singly-charged monoisotopic ion species.
The second challenge is how to explore the search space of potential proteoforms. Because most proteins are post-translationally modified (e.g. proteolytic cleavage, acetylation, etc.), the number of possible proteoforms expands exponentially beyond the number of genes, e.g., over a billion combinatorially possible proteoforms in humans. Popular top-down data analysis tools ProSightPC12,13 and MS-Align+ (recently renamed to TopPIC)14,15 address this challenge using different approaches. ProSightPC restricts the search space to a limited set of proteoforms in a ‘proteome warehouse’, which is a curated collection derived from known PTMs, splice, and single nucleotide variants. While this approach has advantages in confirming known variants and accurately characterizing proteoforms, it has a limitation that it is truly effective only for organisms having a well annotated genome and a well characterized proteome. In contrast, MS-Align+ allows “blind” modifications accounting for any and all PTMs and mutations, and uses the spectral alignment algorithm to efficiently score multiple proteoforms simultaneously. Although this blind search approach is valuable in discovering unknown PTMs and mutations, it may produce substantial amount of false positive proteoform spectrum matches (PrSMs). Recently, another top-down analysis software, named pTop has been developed16. In pTop, the search space is restricted by taking only expected modifications into account, however the current version has no capability to find cleaved or truncated proteoforms.
As a result of these two challenges, proteoform identifications are error prone to mis-localized PTMs, false cleavages or erroneous precursor masses. Thus, it is often necessary for users to manually validate and refine results. Additionally, quantification studies may require researchers to examine the features or Extracted Ion Chromatogram (XIC) of precursor ions of different charge states, or even different regions of the isotopic peak distributions for protein ions. Therefore, visualization tools for top-down proteomics face high demands to assist such data curation12,13,16–18.
We present Informed-Proteomics, a new open source software suite for top-down proteomics analysis. The software package contains a new LC-MS feature finding algorithm (ProMex), a new database search algorithm (MSPathFinder), and an interactive results viewer (LcMsSpectator) (Supplementary Fig. 1). We demonstrate both identification and quantification capabilities of Informed-Proteomics and compare it to other existing tools. The key advantages of Informed-Proteomics over existing software are: high accuracy in LC-MS features detection by ‘smart’ aggregation and summation of features from the same species (which e.g. enhances measurement sensitivity); an efficient algorithm for high-throughput searching proteoforms with combinations of PTMs and truncations by reducing redundancies to minimize the search space; an interactive visualization tool for easy and fast manual validating and refining the results. Informed-Proteomics is an open-source software suite, available on GitHub at https://github.com/PNNL-Comp-Mass-Spec/Informed-Proteomics.
Results
Improvements in the new top-down proteomics data analysis workflow
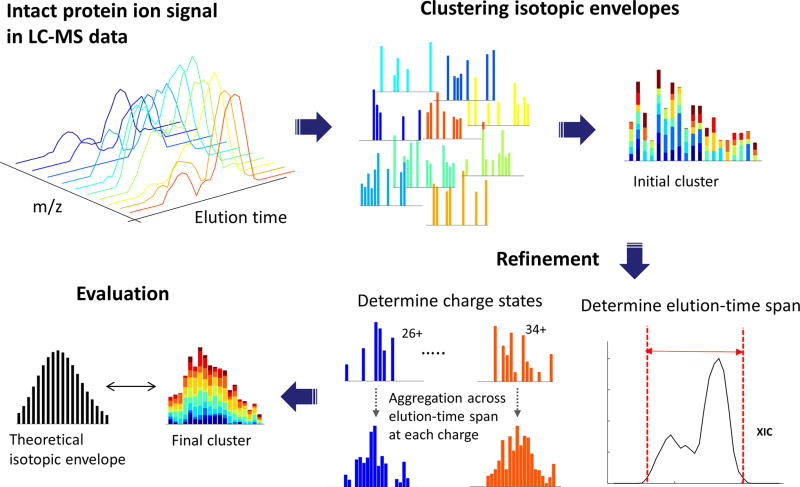
The first component of the Informed-Proteomics software suite, ProMex, finds and characterizes putative proteoforms in LC-MS data. An LC-MS feature represents a group of isotopomer envelopes corresponding to the same putative proteoform ion across all charge states and LC elution times. Due to ions being dispersed widely across LC times, charge states and isotope species, individual isotope envelopes typically have a poor shape compared to expected profiles (Supplementary Fig. 2). ProMex incorporates two key innovations to improve accuracy of feature detection (see Fig. 1). First, ProMex not only aggregates signals across different charge states but also explicitly uses the LC dimension to aggregate features over elution time. Some existing tools (e.g. Xtract in specific commercial implementations) also use the LC dimension by periodically averaging a fixed number of spectra at chromatographic peaks to increase the signal-to-noise ratio. ProMex explicitly looks for all isotopomer envelopes distributed over 3-dimensional LC/MS data and dynamically determines the elution time spans for every candidate mass.
Figure 1. LC-MS feature finding in ProMex.
An LC-MS feature refers to a group of isotopomer envelopes corresponding to the same proteoform species across all charge states and LC elution times. The ProMex algorithm begins with clustering isotopomer envelopes across adjacent time and charge state. The initial cluster is refined to accurately determine its elution-time span and range of charge states. After refinement, ProMex calculates the likelihood that the final cluster is a true LC-MS feature.
Second, rather than examining individual isotopomer envelopes separately, ProMex measures the likelihood of detected LC-MS features based on the aggregated isotopomer envelope. The score is calculated by a likelihood scoring function which takes into account aggregated isotopomer envelope shape, intensity, charge distribution, and the correlation of elution profile at different charge states (see details in Methods). The output of ProMex is a list of LC-MS features defined by monoisotopic mass, range of charge states, elution-time span, abundance, and likelihood scores.
Detected LC-MS features are fed into the database search tool, MSPathFinder, to characterize proteoforms from MS/MS spectra. MSPathFinder operates much like bottom-up proteomics tools, allowing users to specify a set of post-translational modifications and the maximum number of allowable modifications in a sequence. MSPathFinder also provides the statistical significance of PrSMs with E-values computed by the generating function approach19,20, and the false discovery rate (FDR) estimated using the target-decoy approach21.
MSPathFinder efficiently explores the combinatorial proteoform space using a new graph-based approach called the sequence graph (see Fig. 2 for illustration), which allows quickly exploring the vast number of possible proteoforms when considering variable PTMs. There are two important motivations behind the sequence graph. First, because many proteoforms differ only by the location of PTMs, the number of unique elemental compositions is much smaller than the number of proteoforms. Using histone H4 as an extreme example, the number of proteoforms possible when applying 5 modifications (acetylation, methylation, di-methylation of Lys and Arg; tri-methylation of Arg; and phosphorylation of Ser, Thr, and Tyr) is about 50 trillion; but the number of their unique elemental compositions is only 2,344. Second, many fragment compositions are shared by proteoforms with the same composition. Therefore, it is inefficient to score these proteoforms independently. The goal of the sequence graph approach is to effectively remove such redundancies. MSPathFinder, pTop and MS-Align+ all use similar spectral alignment algorithms14,22–25 in a sense that they utilize a parametric dynamic programming algorithm to find the best scoring proteoform in a sequence. However, MSPathFinder uniquely uses node in the sequence graph to represent a composition of Atoms (mostly C, H, N, O, S) rather than a combination of modifications. Since some combinations of modifications have exactly the same atomic compositions (e.g. tri-methylation vs methylation + di-methylation), atom-centric graphs tend to be smaller, which leads to a faster running time in general.
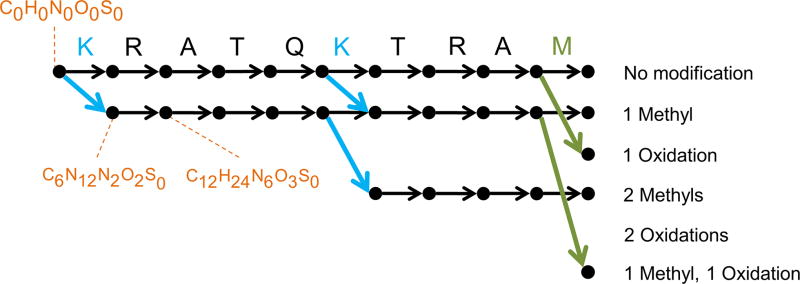
Figure 2. Illustration of the sequence graph for “KRATQKTRAM”.
The sequence graph compactly represents all possible proteoforms of a single atomic composition and facilitates efficient scoring of the search space. In this example, oxidized methionine and methylated lysine are considered as dynamic modifications and up to 2 modifications are allowed per sequence. The graph is constructed from left to right, with the leftmost vertex (source) corresponding to C0H0N0O0S0. The vertically aligned vertices correspond to fragments created by cleaving ith and (i+1)th amino acids. The horizontally aligned vertices represent the fragments with the same modifications. The black, green, and blue edges correspond to unmodified amino acids, oxidized methionine, and methylated lysine, respectively. Each vertex corresponds to a composition and several compositions are shown for illustration. Each of the rightmost vertices (sink) is called a precursor vertex and represents a unique elemental composition of proteoforms with the specified combination of modifications. Thus a path from source to sink represents a proteoform.
MSPathFinder uses a second technique to efficiently explore the vast search space of intact proteoforms. As many proteoforms are enzymatically cleaved or truncated forms of proteins, allowing both N-terminal and C-terminal truncations are necessary to identify the mature processed proteoform, but this substantially increases database search time. In order to reduce the number of possible sequence candidates which serve as query sequences in the search mode of multiple internal cleavages, we implemented a de novo sequencing algorithm to find short amino acid sequences, called sequence tags similar to a previous approach26. Once a protein matches to a sequence tag, MSPathFinder searches multiply cleaved proteoforms of the protein using two sequence graphs toward opposite terminals (Supplementary Figure 3). While this tag-based approach is helpful to restrict the search space significantly, it may fail to find correct proteoforms when a sufficient number of consecutive fragment ions are not detected in MS/MS spectra.
For visualizing and analyzing top-down proteomics data, we created LcMsSpectator as a stand-alone desktop application that is fully integrated with both ProMex and MSPathFinder. This allows maximum data exploration by interacting with both the LC-MS features and also MS/MS identifications. The spectral and chromatographic evidence for the search results are delivered instantly upon completion of the search for comparison with the original identifications. Sequences can also be edited in the application and scored on-the-fly, making it easy to find evidence for proteoforms that were not found in the original database search. LcMsSpectator utilizes a floatable and dockable tabbed-document interface that lets users customize various data grids, spectrum and chromatogram views (see Supplementary Fig. 4). LcMsSpectator supports both automatic and assisted revision of results and identifications (see an example in Supplementary Fig. 5). All of the views and data plots can be exported to high-resolution, publication-ready images.
LC-MS feature detection using ProMex
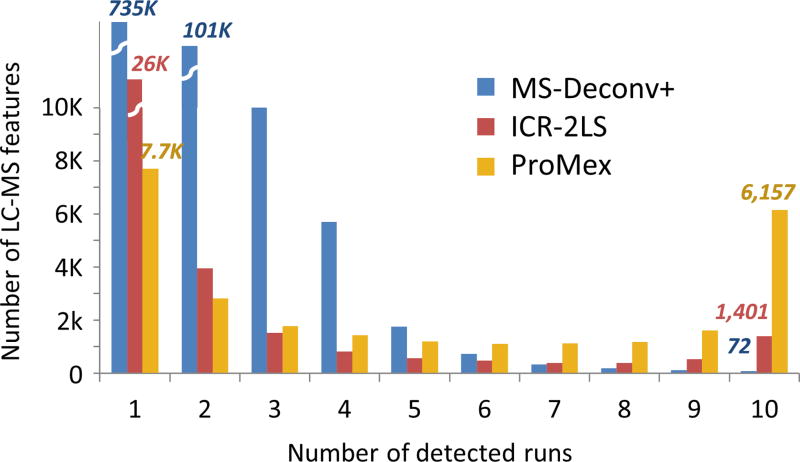
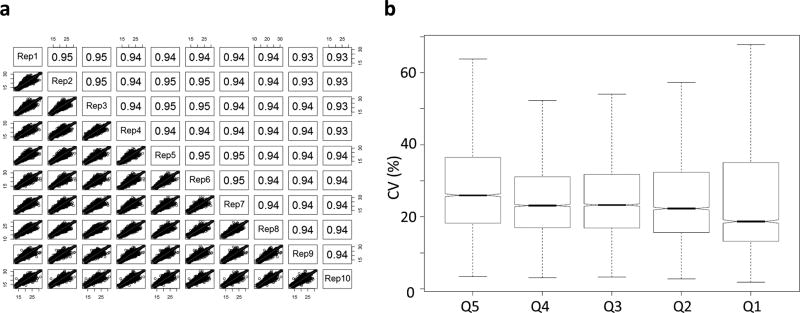
First we assessed the accuracy of our feature detection algorithm, benchmarking ProMex against other MS1 feature detection algorithms including ICR-2LS (http://omics.pnl.gov/software/icr-2ls) and MSDeconv+10,11 (see details in Methods). For this benchmarking test, we created 10 replicate LC-MS/MS data files from ovarian tumor sample. Their average running times of ICR-2LS, MSDeconv+, and ProMex were 180, 23 and 35 minutes, respectively. The metric for accuracy in this test was two-fold. Because they are replicates runs, we anticipate that true features would be present in most files and at similar retention times with similar intensities. As shown in Fig. 3, ProMex had a significantly higher number of features detected in all 10 replicates. MSDeconv+ had an overwhelming number of detected features present in only one or two datasets, pointing to a high variability in the data deconvolution; only 0.04% of features were found in 8 or more datasets. ICR-2LS is an early implementation of the THRASH deconvolution algorithm8. Although it performed substantially better than MSDeconv+, it still had only 6% of features appear in 8 or more datasets. ProMex showed the best performance in reproducible detection of LS-MS features in these replicate datasets, with 34% of features identified in 8 or more datasets. The second metric for determining the accuracy of LC-MS feature detection is the quantitative reproducibility, as this ultimately defines the utility of the methodology for interrogating changes in a system of interest. Fig. 4(a) shows the abundance correlation plots for ProMex identified features for 10 replicate analyses. The high reproducibility of the platform is demonstrated by Pearson correlation coefficients that vary from 0.93–0.95 across all runs. Furthermore, applying our workflow to the ovarian tumor replicates, we were able to achieve coefficients of variation similar to those obtained in label-free bottom-up proteomics27–30 (Fig. 4(b)).
Figure 3. Consistency of LC-MS feature detection.
We examined the reliability of LC-MS feature detection by testing the frequency of feature identification within 10 technical replicate analyses of an ovarian tumor sample. MS-Deconv+, ICR-2LS and ProMex identified 0.04%, 6%, and 34% of features in 8 or more datasets, respectively. In particular, ProMex outperformed other tools in the number of LC-MS features that are detected across all 10 replicates with 6,157 LC-MS features.
Figure 4. Quantitative reproducibility for LC-MS features detected in 10 technical replicate LC-MS analyses.
(a) Correlation plots for 10 replicate injections of ovarian tumor tissue lysate. R-squared values vary from 0.93 – 0.95 demonstrating the reproducibility of the ProMex feature detection algorithm as well as stability of measurement platform, and (b) Coefficient of variance for measured proteoform abundances divided by abundance quintile. Overall median CV for feature abundance was 22.3%.
Proteoform identification using MSPathFinder
Next, we compared the performance of MSPathFinder to that of other top-down database search tools: MS-Align+ (i.e. TopPIC v0.9.1)14,15, pTop v1.216 and ProSightPC v3.013 (see details in Method). We ran each program on the same computer against an ovarian tumor replicate run which contains 3696 MS1 spectra and 4329 MS2 spectra. A human proteome sequence database (UniProt Release 2015_10) which contains 20,209 protein sequences was used for MS-Align+, pTop and MSPathFinder while ProSightPC ran against the annotated human proteoform databases (2014_07 version downloaded from ftp://prosightpc.northwestern.edu). PrSMs identified by MS-Align+, pTop and MSPathFinder were controlled at FDR 1% using the same target and decoy databases. Since pTop v1.2 is not able to search cleaved proteoforms, we compared pTop and MSPathFinder separately, disabling internal cleavages in MSPathFinder. There was no option to run ProSightPC against user-provided target/decoy databases, we therefore used an E-value cut off of 1E-4, which is default cutoff to distinguish good and bad matches in the software.
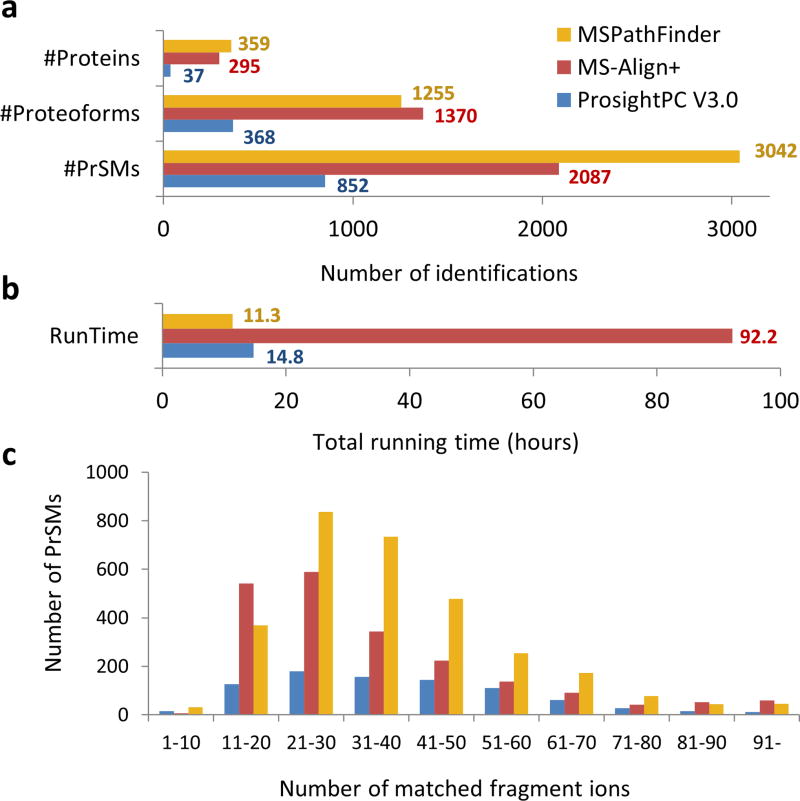
Since each tool explores different regions of the proteoform space, it is difficult to directly compare the results. Moreover, we believe the searches are complimentary and can be used in combination to achieve the best results. MS-Align+ has the greatest search space, and consequently identified the greatest number of unique proteoforms (Fig. 5(a)). ProSightPC has the most restrictive search space and therefore identified the fewest, indicating that even for human samples, the annotation of known proteoforms is often incomplete. MSPathFinder showed dramatically faster run time (11.3 h) than MS-Align+ (92.2 h) and comparable run time with ProSightPC (14.8 h) (Fig. 5(b)). In the comparison with pTop, MSPathFinder found 10–20% more proteins, proteoforms, and protein-spectrum matches (PrSMs) than pTop (Supplementary Table. 1). While the total running time of MSPathFinder was longer than pTop due to the running time of ProMex, MSPathFinder showed a faster running time in database search than pTop. Finally, when we look at the number of annotated peaks in an identified spectrum as a proxy measure for the quality of identifications, MSPathFinder annotates significantly more peaks per spectrum than either TopPIC or ProSightPC (Fig. 5(c)).
Figure 5. Protein identification and characterization results for a human ovarian tumor.
(a) The number of proteins, proteoforms, and protein-spectrum matches (PrSMs) identified by ProsightPC V3.0 (E value ≤ 10−4), MS-Align+ (1% FDR), and MSPathFinder (1% FDR). MSPathFinder achieves more proteins and PrSMs than other tools. Although MS-Align+ achieves 30% less PrSMs than MSPathFinder, it has the maximum number of unique proteoforms due to its unrestrictive PTM search algorithm. (b) Total running time for deconvolution and database search. MS-Align+ took almost four days because its search space much larger than those of other tools. (c) Histogram of the number of matched fragment ions. The majority of PrSMs by MSPathFinder had more matched fragment ions than others.
Label free quantification
As a demonstration of a comparative top-down study, we applied our top-down proteomics workflow for label-free quantification of the human-in-mouse xenograft breast tumor samples31 previously characterized by the Clinical Proteomic Tumor Analysis Consortium32. Two subtypes of breast cancer tumors, basal-like (WHIM2-P32) and luminal B (WHIM16-P33), were analyzed. We created 5 technical replicate analyses for each subtype. First, the LC-MS features detected across all 10 replicates runs were quantified and aligned, and then statistical significance tests were performed to find differentially expressed LC-MS features in the two breast tumor subtypes, WHIM2 and WHIM16 (see Methods). There were a total of 7,300 differentially expressed LC-MS features at adjusted P value of < 0.01 and a fold change of > 1 (Supplementary Fig. 6). Next, we quantified the differential expression of identified proteoforms (Fig. 6). Among a total of 3,207 proteoforms identified in WHIM2 and WHIM6 samples, 1636 proteoforms were found to be differentially expressed with adjusted P value of < 0.01 and fold change of > 2. Recently, an integrated approach of bottom-up and top-down proteomics to detect differentially expressed protein and proteoforms was reported for this same tumor comparison33. In both LC-MS feature- and proteoform-level analysis, our top-down proteomics workflow found 10 times more differentially expressed entities than the approach described in the article. Furthermore, we achieved this characterization using only 30 hours of instrument time as compared to 200 hours reported in the literature.
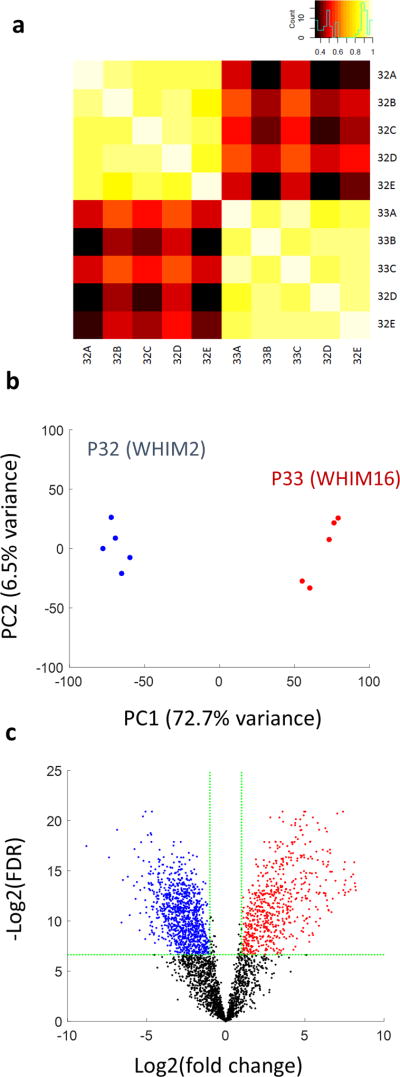
Figure 6. Differentially expressed proteoforms in CompRef breast tumor sample.
Two breast cancer xenograft tumors representing basil-like (P32) and luminal B (P33) subtypes were compared to identify differentially expressed proteoforms. Five technical replicate LC-MS/MS analyses were performed for each tumor. (a) Pearson correlation plot (b) PCA plot and (c) volcano plot. 622 protoeoforms are up-regulated in P32 (WHIM2) while 1014 proteoforms are up-regulated in P33 (WHIM16) at 1% FDR and fold change > 2.
In order to demonstrate the effectiveness of our top-down data analysis pipeline, we analyzed the same dataset used in one of the studies in the article with Informed-Proteomics. Using the same statistical model, we found total 412 differentially expressed proteoforms mapping to 280 proteins with adjusted P value of < 0.01 and absolute log2 fold change of > 1. In comparison to the recent article, our analysis pipeline found 2.7 and 2.4 times more differentially expressed proteoforms and proteins, respectively (Supplementary Fig. 7).
Discussion
We present an open source software suite, Informed-Proteomics, for top-down proteomics analysis. The software suite includes an LC-MS feature finding algorithm (ProMex), a new database search algorithm (MSPathFinder), and an interactive results viewer (LcMsSpectator). Our software suite is designed for a sensitive and comprehensive high throughput analysis of complex mixtures of intact proteins. ProMex aggregates signals not only across different charge states, but also over LC time such that it measures the likelihood of detected LC-MS features based on the aggregated isotopomer envelope. ProMex relies on isotopically resolved peaks and is designed for high-resolution LC-MS data. Therefore, it does not currently work for data with only a charge envelope or data without an LC separation. We demonstrated that ProMex accurately recovers more common features and less uncommon (irreproducible) features across multiple replicate runs than other existing algorithms. We also showed that our database search algorithm efficiently explores the combinatorial proteoform space using a new graph-based approach called the sequence graph. MSPathFinder operates in a similar manner as most common bottom-up proteomics algorithms, requiring users to enumerate specific post-translational modifications of interest. It does not discover unknown modifications, which can be done with complementary algorithms for open PTM search. As an application of comparative top-down proteomics, we showed how ProMex and MSPathFinder find differently expressed LC-MS features and proteoforms from breast cancer samples.
Online Methods
Intact protein extraction and preparation for LC-MS/MS analysis
Ovarian tumor sample
Ovarian tissue used in this manuscript was from a pool of 5 female patients; the samples were collected with the oversight of the Institutional Review Board at Oregon Health and Science University and the patients gave informed consent. An approximately 20 mg portion of fresh frozen tissue was taken to create a pooled sample. Tissue aliquots were homogenized with a pellet pestle in 1 mL of homogenization buffer (8 M Urea, 50 mM Ammonium bicarbonate, 1 mM PMSF, and 1% Sigma phosphatase inhibitor cocktail II and III). The resulting homogenate is then incubated at 37 °C for 30 min to facilitate protein extraction, and spun for 10 min to pellet insoluble debris. All centrifugation steps were carried out at 4 °C, to further limit potential enzymatic activity, and 15000 rpm. The supernatant is then transferred to an Amicon ultra 100K MWCO filter (EMD Millipore) pre-rinsed with 500 uL homogenization buffer. Samples were then centrifuged for 30 min obtain minimum volume. A 450 uL aliquot of homogenization buffer was added to the filter and spun for an additional 30 min to maximize protein recovery. The filtrate was then transferred to an Amicon ultra 10K MWCO filter and spun for 40 min to obtain the minimum volume. Buffer exchange was achieved using 3 washes with 450 uL of buffer A (3% Acetonitrile, 0.2% formic acid in MilliQ water). The protein concentration was determined using a Coomassie assay (Thermo Fisher). The final protein concentration was adjusted to 0.5 ug/uL for analysis. Ten replicate LC-MS/MS datasets were acquired.
Breast tumor xenograft sample
We created top-down LC-MS/MS datasets for two subtypes of breast cancer tumors: basal-like (WHIM2-P32) and luminal B (WHIM16-P33) (see ref 32). For each subtype, six process replicates LC-MS/MS runs were created. Tumors were cryopulverized, distributed into six aliquots for each tumor and stored −80 °C until use. The six aliquots were processed independently for each tumor as described above. All samples were block-order randomized. Each sample was then analyzed by a single 180 min LC-MS/MS run. The first injection from each tumor was used to passivate the new LC column, and these files were not included in later analyses.
LC-MS/MS analysis
A dual pump Waters nanoACQUITY™ UPLC system (Millford, MA) in combination with an Velos Orbitrap Elite mass spectrometer (Thermo Fisher, San Jose, CA) was used for these analyses. A 5 µL sample injection was loaded on a solid phase extraction (SPE) column for rapid trapping and desalting prior to separation. The analytical column was prepared in-house by slurry packing 3-µm diameter C2 stationary phase (Separation Methods Technology, Newark, DE) into a 50-cm length of 360 µm o.d. × 100 µm i.d. fused silica capillary column (Polymicro Technologies Inc., Phoenix, AZ). The SPE column (360 µm o.d. × 150 µm i.d.) of 5 cm length was similarly prepared. Mobile phases consisted of 0.2% formic acid in water (A) and 0.2% formic acid in acetonitrile (B). Sample was loaded for 30 minutes on the SPE column and then separated by the analytical column using a 190 minute gradient from 99% A to 35% A in 180 minutes at a flow rate of 0.3 uL/min. The LC column was interfaced with the mass spectrometer using a home-made nano-electrospray ionization source with a chemically etched 150 µm o.d. × 20 µm i.d. fused silica emitter. A spray voltage of 2.3 kV and an ion transfer tube temperature of 325°C were used for ionization and de-solvation. Precursor spectra were acquired from m/z 500 to 2000 at a resolution of 240,000. Data-dependent product spectra of the top 4 ions were isolated in a 4 Dalton window and subjected to CID and HCD fragmentation modes at normalized collision energies of 35% and 30%, respectively. All product ions were detected in the Orbitrap at a resolution of 120,000.
Data format for LC-MS/MS spectrometry
For fast data access, MSPathFinder and ProMex use an internal file format for LC-MS data, called a pbf file. This file stores LC-MS data as a collection of three-dimensional peaks: scan number, m/z and intensity. To support quick retrieval of both spectra and chromatograms, the PBF format indexes peaks in two ways: (1) spectrum-centric way - get all the peaks for a certain scan number, and (2) chromatogram-centric way - get all the peaks within a specified m/z range.
ProMex: LC-MS feature extraction
ProMex was developed to detect isotopomer envelopes of intact protein ions and determine their monoisotopic masses and abundances. The ProMex algorithm takes a range of monoisotopic mass and a mass tolerance as an input, and then outputs a collection of LC-MS features, each of which is specified by monoisotopic mass, charge states, elution-time span and abundance. The basic idea is that an individual isotopomer envelope of one charge state in one spectrum has poor ions statistics, especially as molecular weight increases (see Supplementary Fig. 2). Therefore, we evaluate an isotopic profile grouped across time and charge. The set of peaks attributed to a single proteoform species (across time and charge state) is referred to as an LC-MS feature. The process of determining which peaks belong to the same LC-MS feature is shown graphically in Fig. 1 and described in the subsequent paragraphs.
To identify which peaks in LC-MS data should be grouped, a list of potential masses is created using the user specified mass range and tolerance. For each potential monoisotopic mass M (Da), a theoretical isotopomer envelope EM is generated from Averagine model34; then using the input charge range various m/z values are calculated for EM. ProMex then scans all MS1 spectra to identify peaks corresponding to these isotopomer envelopes. The collected peaks are grouped by their charge states and elution times, and thus an observed isotopomer envelope at charge state ci and elution time tj is denoted as Eij
The second step is to cluster isotopomer envelopes indicating the same proteoform species across charge states and LC elution time. ProMex gathers envelope peaks in adjacent charges and elution times using a greedy algorithm. It starts with seed isotopomer envelopes selected based on their similarity scores against EM and statistical significances. Here the similarity score S(E1, E2) between two envelopes, E1 and E2 is computed by the Pearson correlation. The statistical significance is determined by Wilcoxon rank sum test and hypergeometric test, as previously described35. Both tests are performed within a local range (5 m/z) of the spectrum encompassing the seed envelope. Seed envelopes must have p-values less than 0.01 for both tests and similarity scores larger than specified thresholds. The increasing number of charge states and the increasing size of isotopomer envelope lower the chance to observe highly similar isotopomer envelopes to the theoretical one. Thus, we use different thresholds depending on the mass M: 0.7 for M < 10,000 Da, 0.6 for 10,000 < M < 15,000, 0.5 for 15,000 < M < 30,000, 0.4 for 30,000 < M.
The clustering process starts with the seed envelope having the highest similarity score among the seed set. The greedy algorithm iteratively explores observed envelopes in adjacent charge states and elution times. Adjacent envelopes are added to the cluster if they enhance the similarity between aggregated isotopomer envelope in the cluster against EM. When there is no adjacent isotopomer envelope improving the cluster, it stops exploration. This process continues until all seed envelopes are assigned to clusters.
Detected clusters are refined to accurately determine their elution-time spans and ranges of charge states. The elution-time span is determined based on elution profile (EP). The EP is constructed by peaks in the clustered isotopomer envelopes, and smoothed by Savitzky-Golay filter using nine adjacent points with quadratic polynomial. The first and last elution times having intensities equal or greater than 1% of the apex intensity are set to elution start (tmin) and end time (tmax), respectively. To determine the range of charge states, at each possible charge state ci, it examines not only individual isotope envelopes Eij in the elution-time span (tmin ≤ j ≤ tmax) but also aggregated isotopic envelop Ei over the span. If either any single Eij or Ei has similarity score higher than 0.7, the charge state is included into the cluster. Thus, the minimum (cmin) and maximum (cmax) charge states define the range of charge states. The final monoisotopic mass of LC-MS feature is determined by selecting the median value from all the clustered isotopomer envelopes.
The abundance of LC-MS features is measured by the area under EP. In order to avoid outlier peaks due to signal interference or noise, it only includes peaks in isotopomer envelope Eij where S(Eij, EM) > min(0.7, median({S(Eij, EM)|cmin ≤ i ≤ cmax, tmin ≤ j ≤ tmax}). The area under smoothed EP is calculated and set to the abundance of LC-MS feature.
The quality of each feature is evaluated by a likelihood-ratio scoring function. Features that fail the likelihood test (i.e. are not distinguishable from random) are rejected and deleted. We devised a Bayesian network that models LC-MS features to determine the probability of observing aggregated isotopomer envelopes Ei given mass M (Supplementary Fig. 8). A series of isotopomer envelopes detected in the elution-time span at a charge are described by four parameters, Ai, Si, Ii and Xi. Here Ai is the ratio of abundance at charge ci to total abundance, and Si is the similarity score S(Ei, EM) of aggregated isotopomer envelope Ei. At each spectrum, the intensity of isotopic peaks is scaled by dividing them the highest intensity in a window of width 5 m/z around the isotope envelope. Ii is the sum of scaled intensities of the most abundant isotopic peaks within the elution-time span. Xi is elution profile score which is the average Pearson’s correlation coefficient of EP at charge ci against EPs at other charge states. Thus, the likelihood scoring function can be represented as
where Pobs is the probability of a particular state (Ci, Ai, Si, Ii Xi|M) observed in a sample of known LC-MS features and Pnull is the probability of the same state in a null hypothesis model where peaks are randomly shuffled over 3D LC-MS space.
Considering conditional dependencies of parameters as defined in Supplementary Fig. 8 and applying Bayes’ theorem, the likelihood scoring function can be rewritten as
The detected LC-MS features often overlap and share peaks with each other because a cluster of observed peaks can be well matched to different theoretical isotopomer envelopes (see Supplementary Fig. 9). To eliminate redundant, false LC-MS features, ProMex selects only the best scoring features iteratively and removes them from the LC-MS data. For this, ProMex constructs an undirected acyclic graph where each vertex represents an LC-MS feature. Two vertices are connected by an edge if they share peaks in their collected isotopomer envelopes. Vertices are grouped such that two vertices in a group are connected to each other by paths. In each group, the best scoring LC-MS feature is selected and peaks associated with the feature are removed from the LC-MS data. If there are LC-MS features with ±1 Da differ from the best scoring feature, they are selected together to maximize the chance of identifying correct proteoforms. Whenever peaks are removed from the LC-MS data, other remaining features are rescored. This process is repeated until the best score in the group is less than a certain likelihood score cutoff.
MSPathFinder: proteoform identification
MSPathFinder takes an LC-MS feature file generated by ProMex, a protein FASTA database, and a set of search modifications as an input and outputs PrSMs with E-values. A search modification is defined as a pair consisting of a PTM and a target amino acid. The maximum number of allowable modifications is also given as input. For each sequence present in the protein database, it constructs a sequence graph (described below) and scores proteoforms against MS/MS spectra through graph searching. The statistical significance of individual PrSMs (e.g. E-values) is also evaluated. Lastly, it estimates the false discovery rate.
Enumerating protein substrings
MSPathFinder supports 3 search modes depending on the number of internal cleavages allowed. The search mode 2, similar to the non-tolerable termini (NTT) 2 in bottom-up proteomics, does not allow any internal cleavage except the single amino acid cleavage at the N-terminus. The search mod 1, similar to NTT 1, additionally allows single internal cleavage and the search mod 0 (similar to NTT=0) allows multiple internal cleavages. The numbers of sequences to be searched are different depending on the search mode. Also, the lowest and highest masses of detected LC-MS features also provide lower and upper bounds in sequence lengths.
MS/MS spectra deconvolution
MSPathFinder uses a fitting method similar to THRASH algorithm8 to deconvolute MS/MS spectra. The deconvolution algorithm moves a window of a certain m/z width along the peaks (here, 2.2 m/z was used). The most intense peak in the window is selected, and a few number of average mass are calculated using the observed m/z for a certain range of charge states (here, charge states of 1–20+ were used). For each average mass, a theoretical isotopomer envelope is generated from Averagine model34. Then, it identifies observed isotope peaks corresponding to the theoretical isotope envelope. Pearson’s correlation coefficient between observed and theoretical isotopomer envelopes is computed. If the correlation coefficient is higher than a certain threshold (here, 0.7 was used), the observed isotopomer envelope is converted to a deconvoluted peak defined by monoisotopic mass, charge state, and intensity.
Sequence graph
The sequence graph is a directed acyclic graph (DAG) that represents all possible PTM-modified forms of a protein sequence (see Fig. 2). Each vertex of the sequence graph represents a unique fragment and corresponds to an elemental composition. Two vertices are connected by an edge if their difference in compositions equals to a composition of an amino acid and optionally a PTM. For example, C6H12N2O2S0 and C12H24N6O3S0 are connected by an edge representing Arg (C6H12N4O1S0). In the sequence graph, the leftmost vertex (called the source) represents the “zero” fragment with a composition C0H0N0O0S0 and each of the rightmost vertices (called a precursor vertex) represents proteoforms with the same modifications. Each path in the graph corresponds to a proteoform.
Once the sequence graph is constructed, MSPathFinder selects a precursor vertex νp one by one, and repeats the following procedure. It considers a subgraph from the source to νp. This subgraph represents the proteoforms with the same composition and each internal vertex represents a fragment of these proteoforms. For the precursor mass of νp, MS/MS spectra are retrieved from the LC-MS look-up table. For each recruited MS/MS spectrum and each internal node ν, it finds evidence of the ions generated by a fragment with a composition ν and assigns a score to ν. Edge scores are also assigned as necessary (e.g. consecutive fragment ion score). The score of a path is defined as the sum of scores of vertices and edges in the corresponding path. MSPathFinder finds the best scoring proteoform by backtracking the sequence graph.
MSPathFinder Scoring
We designed a scoring function, MSPathScore(P, S) to evaluate a PrSM of a proteoform P and a spectrum S. The MSPathScore utilizes five characteristics of matched fragment ion peaks: intensity, isotopomer envelope shape, mass measurement error, existence of complementary fragment ion peak, and existence of consecutive fragment ion peaks, which can be written as:
where α and β are sets of prefix and suffix fragment ion peak matches, respectively. Ii, Di, and Ei are normalized intensity, isotopomer envelope similarity score, and mass error of matched fragment ion i. The normalized intensity is calculated by dividing the peak’s intensity by the highest intensity in the spectrum. The envelope similarity is determined by Pearson’s correlation coefficient between observed and theoretical isotopomer envelopes. The mass error is measured in ppm. IsComple(i, j) is an indicator function to denote whether the fragment ion pair (i, j) is a complementary fragment ion pair. IsConse(i, j) is also an indicator function describing the ions (i, j) are consecutive fragment ions. The weight parameters in MSPathScore were determined by a logistic regression method with a training set of 30,000 PrSMs. The training set was obtained by scoring PrSMs as the number of matched fragment ion peaks.
Sequence tag based search
MSPathFinder uses sequence tags to filter the search space of multiply cleaved protein sequences. Sequence tags are short amino acid sequences that are found by combining consecutive fragment ions in protein sequences26. MSPathFinder generates all possible sequence tags with a minimum length, and finds multiply cleaved protein sequences containing the sequence tags. Here, a minimum length of 5 residues is chosen as it gives a good balance between the number of identifications and the size of search space. Given a protein sequence matched to a sequence tag, two sequence graphs originating from the both ends of sequence tags are generated toward opposite directions as shown in Supplementary Fig. 3. The flanking mass of sequence tags and the mass of LC-MS features are used to constrain candidate proteoforms to be searched.
Statistical significance of Protein-Spectrum Match (PrSM)
The statistical significance such as P-value or E-value is estimated by the generating function approach as previously described19,20. The implementation of generating function for bottom-up proteomics is not directly applicable to top-down proteomics, not only because of large masses of intact proteins but also because of increased mass accuracy and resolution. In order to minimize the number of integer masses to be considered in the generating function while accommodating the high mass accuracy, we discretize the real mass space with a window of a constant mass tolerance (e.g. 8 parts per million). In addition, masses within mass regions that cannot be reached by combinations of amino acid with allowable PTMs masses are removed.
Estimating FDRs
The false discovery rate (FDR) is estimated using the target-decoy approach21, where a decoy database was constructed by reversing the protein sequences and applying 3 amino acid mutations at random positions.
LcMsSpectator: visualization and refinement tool
LcMsSpectator is a Windows desktop application facilitates to visualization and refinement of top-down proteomics analysis results reported by ProMex and MSPathFinder and features rich, interactive spectral and chromatographic data plots as shown Supplementary Fig. 4. All of the views and data plots can be exported to high-resolution, publication-ready images. LcMsSpectator supports the community standards for both spectrum data (mzML36) and spectrum annotation (mzIdentML37). See https://github.com/PNNL-Comp-Mass-Spec/LCMS-Spectator for details.
Software evaluation
All the experiments were performed on a Windows computer with a 3.5 GHz CPU (Intel Xeon E3-1270) and 32 GB memory.
Comparison of LC-MS feature detection algorithms
We ran ProMex, MS-Deconv+, and ICR-2LS against total 10 replicate runs. MS-Deconv+ and ICR-2LS reported monoisotopic masses, charges, elution times and intensities for each MS1 spectrum. We clustered these deconvolution results into LC-MS features if two monoisotopic masses are within the mass tolerance and elution time window. The abundance was computed by summing intensities of cluster members. We repeated this clustering procedure to group LC-MS features detected from multiple replicate runs. Here we used 10 ppm mass tolerance and 1 minute elution time window, respectively. Parameter settings used in ProMex, MS-Deconv+, and ICR-2LS are described in Supplementary Table 2.
Comparison of proteoform identification algorithms
We benchmarked MSPathFinder against TopPIC v0.9.1 (available at http://proteomics.informatics.iupui.edu/software/toppic/)14, ProSightPC v3.013, and pTop v1.2 (available at http://pfind.net/software/pTop/index.html)16. For MSPathFinder, pTop and MS-Align+, used a fasta file of human and mouse from UniProt database for target database (2011_12 version). The same decoy database was used for three tools and PrSMs were collected at 1% FDR. For MS-Align+, the raw spectra files were converted to .msalign file format using msconvert tool in ProteoWizard software package38 and MS-Deconv10 as specified in TopPIC manual. Since pTop v1.2 is not able to search cleaved proteoforms, we compared pTop and MSPathFinder separately, disabling internal cleavages in MSPathFinder. For ProSightPC, the annotated mouse and human proteoform databases (2014_07 version) were downloaded from ftp://prosightpc.northwestern.edu. We tested ProSightPC based on the biomarker discovery search mode with mass tolerance of 10 ppm for precursor and fragment ions. We tried to use shuffled database search option to calculate FDR at ProsightPC. But there were very few PrSMs for shuffled sequences even though E-value cutoff increased to 10. Also, as the downloaded databases are encoded in binary format, we were not able to apply our target-decoy approach. Thus PrSMs were collected with E-value cutoff of 1E-4 (default cutoff for good matches in ProSightPC). Parameter settings used in MSPathFinder, MS-Align+, ProSightPC, and pTop are described in Supplementary Table 3.
Statistical analysis for label-free quantification
LC-MS features are not always detected in all replicate runs. For missing LC-MS features, we integrated background signal intensities in the appropriate elution-time spectra. Here we assumed that median intensity in each spectrum is equal to background signal intensity. Grouped LC-MS features were associated with proteoform identification results by MSPathFinder. Each group of LC-MS features is assumed to be a single proteoform species. If there are multiple different proteoforms matched to a LC-MS feature group, the best scoring proteoform (i.e. the lowest E-value PrSM) was selected as a representative proteoform of the LC-MS feature group. Informed-Proteomics does not provide a separate tool for this post-processing step, but implemented source codes for LC-MS feature grouping was included in the package.
As a pre-processing step for normalization, we normalized the abundance values by equalizing the median of abundances across replicate runs. Conventional ANOVA analysis was performed and p-values were adjusted by Benjamini–Hochberg (BH) procedure. All statistical analyses including ANOVA analysis and principal component analysis (PCA) were performed within MATLAB 2014b (The MathWorks, Inc., Natick, MA).
Supplementary Material
Acknowledgments
Portions of this work was supported by the NIH National Institute of General Medical Sciences grant GM103493 (RDS), the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC) grant U24CA160019 (RDS), the National Institute of Allergy and Infectious Diseases NIH/DHHS through interagency agreement Y1-A1-8401-01 (Joshua Adkins, PNNL), the U.S. Department of Energy (DOE) Office of Science and Office of Biological and Environmental Research, under the Pan-omics program (RDS). LPT, NT, MZ and JBS were supported as part of the "High Resolution and Mass Accuracy Capability" development project at the Environmental Molecular Science Laboratory (EMSL), a U.S. DOE national scientific user facility at Pacific Northwest National Laboratory (PNNL) in Richland, WA. Battelle operates PNNL for the DOE under contract DE-AC05-76RLO01830.
Footnotes
Author contributions
JP, PDP, SHP, SK designed and executed the study. JP, CW, JM, GMF, BCG, SK implemented algorithms in software. TL contributed samples. PDP, YS, AKS, RJM performed LC-MS/MS experiments. JP, PDP, JBS, VAP, MZ, TL, NT analyzed data. LPT, RDS provided technical leadership and oversight. JP, PDP, SK contributed to writing the manuscript with input from all authors.
Competing financial interests
The authors declare no competing financial interests.
Software availability
MSPathFinder’s home page is https://omics.pnl.gov/software/mspathfinder. All source codes were written in Microsoft C# with .NET framework 4.5 and are available at GitHub, https://github.com/PNNL-Comp-Mass-Spec/Informed-Proteomics and https://github.com/PNNL-Comp-Mass-Spec/LCMS-Spectator. Each repository has a readme and wiki to describe installation and usage. Binary executables and installers are available at: https://github.com/PNNL-Comp-Mass-Spec/Informed-Proteomics/releases, https://github.com/PNNL-Comp-Mass-Spec/LCMS-Spectator/releases. A tutorial is available at https://github.com/PNNL-Comp-Mass-Spec/Informed-Proteomics/wiki/MSPathFinder-Tutorial.
Data Availability
All datasets are submitted to the MassIVE proteomics repository under identifier: MSV000080257.
References
- 1.Garcia BA. What Does the Future Hold for Top Down Mass Spectrometry? J. Am. Soc. Mass Spectrom. 2010;21:193–202. doi: 10.1016/j.jasms.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nat. Methods. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith LM, et al. Proteoform: a single term describing protein complexity. Nat. Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Z, Wu S, Stenoien DL, Paša-Tolić L. High-Throughput Proteomics. Annu. Rev. Anal. Chem. 2014;7:427–454. doi: 10.1146/annurev-anchem-071213-020216. [DOI] [PubMed] [Google Scholar]
- 5.Tran JC, et al. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chait BT, et al. Chemistry. Mass spectrometry: bottom-up or top-down? Science. 2006;314:65–6. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- 7.Lanucara F, Eyers CE. Top-down mass spectrometry for the analysis of combinatorial post-translational modifications. Mass Spectrom. Rev. 2013;32:27–42. doi: 10.1002/mas.21348. [DOI] [PubMed] [Google Scholar]
- 8.Horn DM, Zubarev RA, McLafferty FW. Automated reduction and interpretation of high resolution electrospray mass spectra of large molecules. J. Am. Soc. Mass Spectrom. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 9.Zabrouskov V, Senko MW, Du Y, Leduc RD, Kelleher NL. New and Automated MSn Approaches for Top-Down Identification of Modified Proteins. J. Am. Soc. Mass Spectrom. 2005;16:2027–2038. doi: 10.1016/j.jasms.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, et al. Deconvolution and Database Search of Complex Tandem Mass Spectra of Intact Proteins: A COMBINATORIAL APPROACH. Mol. Cell. Proteomics. 2010;9:2772–2782. doi: 10.1074/mcp.M110.002766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kou Q, et al. A new scoring function for top-down spectral deconvolution. BMC Genomics. 2014;15:1140. doi: 10.1186/1471-2164-15-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LeDuc RD, et al. ProSight PTM: an integrated environment for protein identification and characterization by top-down mass spectrometry. Nucleic Acids Res. 2004;32:W340–W345. doi: 10.1093/nar/gkh447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zamdborg L, et al. ProSight PTM 2.0: improved protein identification and characterization for top down mass spectrometry. Nucleic Acids Res. 2007;35:W701–6. doi: 10.1093/nar/gkm371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, et al. Protein Identification Using Top-Down Spectra. Mol. Cell. Proteomics. 2012;11:M111.008524–M111.008524. doi: 10.1074/mcp.M111.008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kou Q, Xun L, Liu X. TopPIC: A software tool for top-down mass spectrometry-based proteoform identification and characterization. Bioinformatics. 2016:btw398. doi: 10.1093/bioinformatics/btw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun R-X, et al. pTop 1.0: A High-Accuracy and High-Efficiency Search Engine for Intact Protein Identification. Anal. Chem. 2016;88:3082–3090. doi: 10.1021/acs.analchem.5b03963. [DOI] [PubMed] [Google Scholar]
- 17.Cai W, et al. MASH Suite Pro: A Comprehensive Software Tool for Top-Down Proteomics. Mol. Cell. Proteomics. 2016;15:703–714. doi: 10.1074/mcp.O115.054387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guner H, et al. MASH Suite: A User-Friendly and Versatile Software Interface for High-Resolution Mass Spectrometry Data Interpretation and Visualization. J. Am. Soc. Mass Spectrom. 2014;25:464–470. doi: 10.1007/s13361-013-0789-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S, Gupta N, Pevzner PA. Spectral Probabilities and Generating Functions of Tandem Mass Spectra: A Strike against Decoy Databases. J. Proteome Res. 2008;7:3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, Pevzner PA. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 22.Pevzner PA, Dančík V, Tang CL. Mutation-Tolerant Protein Identification by Mass Spectrometry. J. Comput. Biol. 2000;7:777–787. doi: 10.1089/10665270050514927. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, et al. Identification of ultramodified proteins using top-down tandem mass spectra. J. Proteome Res. 2013;12:5830–5838. doi: 10.1021/pr400849y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank AM, Pesavento JJ, Mizzen CA, Kelleher NL, Pevzner PA. Interpreting top-down mass spectra using spectral alignment. Anal. Chem. 2008;80:2499–2505. doi: 10.1021/ac702324u. [DOI] [PubMed] [Google Scholar]
- 25.Tsur D, Tanner S, Zandi E, Bafna V, Pevzner PA. Identification of post-translational modifications by blind search of mass spectra. Nat. Biotechnol. 2005;23:1562–1567. doi: 10.1038/nbt1168. [DOI] [PubMed] [Google Scholar]
- 26.Frank A, Tanner S, Bafna V, Pevzner P. Peptide Sequence Tags for Fast Database Search in Mass-Spectrometry. J. Proteome Res. 2005;4:1287–1295. doi: 10.1021/pr050011x. [DOI] [PubMed] [Google Scholar]
- 27.Domon B, Aebersold R. Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 2010;28:710–721. doi: 10.1038/nbt.1661. [DOI] [PubMed] [Google Scholar]
- 28.Nagaraj N, Mann M. Quantitative Analysis of the Intra- and Inter-Individual Variability of the Normal Urinary Proteome. J. Proteome Res. 2011;10:637–645. doi: 10.1021/pr100835s. [DOI] [PubMed] [Google Scholar]
- 29.Zhu W, Smith JW, Huang C-M. Mass Spectrometry-Based Label-Free Quantitative Proteomics. J. Biomed. Biotechnol. 2010;2010:1–6. doi: 10.1155/2010/840518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian W-J, Jacobs JM, Liu T, Camp DG, Smith RD. Advances and Challenges in Liquid Chromatography-Mass Spectrometry-based Proteomics Profiling for Clinical Applications. Mol. Cell. Proteomics. 2006;5:1727–1744. doi: 10.1074/mcp.M600162-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S, et al. Endocrine-Therapy-Resistant ESR1 Variants Revealed by Genomic Characterization of Breast-Cancer-Derived Xenografts. Cell Rep. 2013;4:1116–1130. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabb DL, et al. Reproducibility of Differential Proteomic Technologies in CPTAC Fractionated Xenografts. J. Proteome Res. 2016;15:691–706. doi: 10.1021/acs.jproteome.5b00859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ntai I, et al. Integrated Bottom-Up and Top-Down Proteomics of Patient-Derived Breast Tumor Xenografts. Mol. Cell. Proteomics. 2016;15:45–56. doi: 10.1074/mcp.M114.047480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Senko MW, Beu SC, McLaffertycor FW. Determination of monoisotopic masses and ion populations for large biomolecules from resolved isotopic distributions. J. Am. Soc. Mass Spectrom. 1995;6:229–233. doi: 10.1016/1044-0305(95)00017-8. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, et al. JUMP: A Tag-based Database Search Tool for Peptide Identification with High Sensitivity and Accuracy. Mol. Cell. Proteomics. 2014;13:3663–3673. doi: 10.1074/mcp.O114.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martens L, et al. mzML--a community standard for mass spectrometry data. Mol. Cell. Proteomics. 2011;10:R110.000133. doi: 10.1074/mcp.R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones AR, et al. The mzIdentML Data Standard for Mass Spectrometry-Based Proteomics Results. Mol. Cell. Proteomics. 2012;11:M111.014381–M111.014381. doi: 10.1074/mcp.M111.014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chambers MC, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.