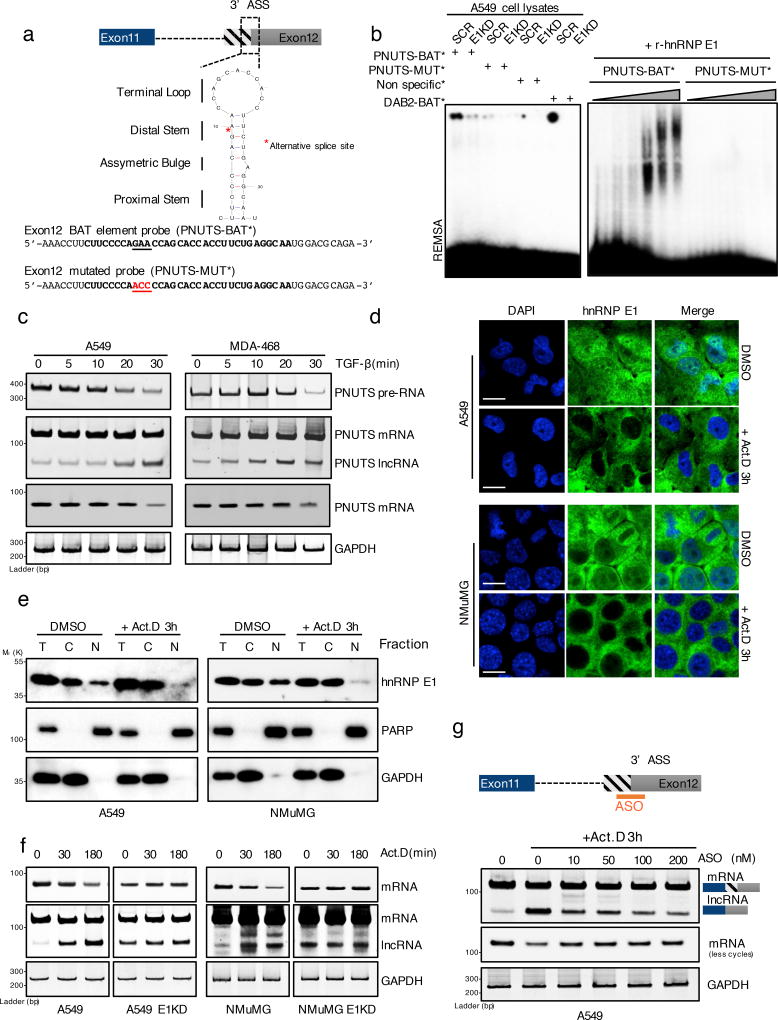
Figure 2. hnRNP E1 protein prevents PNUTS alternative splicing by its specific binding to a BAT structural element.
(a) Secondary structure of the human PNUTS alternative splicing site as predicted by the Mfold algorithm (ΔG= −3.90 kcal.mol−1). The underlined nucleotides colored in red represent the mutant probe used for the REMSA experiment in (b) and (c). The red asterisk represents the exact alternative splicing site leading to the lncRNA-PNUTS isoform generation. (ASS: Alternative Splicing Site)
(b) (Left) RNA electromobility shift assay (REMSA) experiment using either wild-type [PNUTS-BAT] or mutated [PNUTS-MUT] α-32P-labelled PNUTS alternative splicing site probes combined with control (SCR) or hnRNP E1 knockdown (E1KD) A549 cell lysates. The [PNUTS-MUT] probe was mutated by a nucleotide substitution to destroy its secondary structure. [Non specific] and [DAB2-BAT] α-32P-labelled probes were used as negative and positive controls respectively. [DAB2-BAT] corresponds to the BAT-sequence located on the Dab2–3'UTR already described to bind to hnRNP E1. (Right) REMSA using a combination of [PNUTS-BAT] or mutated [PNUTS-MUT] α-32P-labelled probes with increasing concentration of recombinant hnRNP E1 protein purified from e. coli bacteria.
(c) Time course experiment using RT-PCR analysis of PNUTS gene processing after addition of 5ng.mL−1 of TGFβ.
(d) Confocal microscopy imaging of the hnRNP E1 nucleocytoplasmic shuttling by addition of 5µg.mL−1 of Actinomycin D for 3h in A549 and NMuMG cells cultures. Scale bar: 10µM
(e) Characterization of the nucleocytoplasmic transportation of hnRNP E1 following Act.D treatment by using cell fractionation and subsequent western-blot analysis of hnRNP E1 expression. To check the fractions purity, GAPDH and PARP were used as cytoplasmic and nuclear compartment markers respectively.
(f) Time course experiment using RT-PCR analysis of PNUTS predicted-lncRNA alternative splicing activation upon addition of 5µg.mL−1 of Act.D in control (CTRL) or hnRNP E1 silenced (E1KD) A549 and NMuMG cells.
(g) Inhibition of alternative splicing induced by Act.D in A549 cells using and antisense oligonucleotide (ASO) targeting the alternative splicing site of PNUTS.
GAPDH was used as a loading control. Unprocessed original scans of blots are shown in Supplementary Fig. 7