Figure 3. PNUTS alternative splicing product is non-coding and interacts with miR-205.
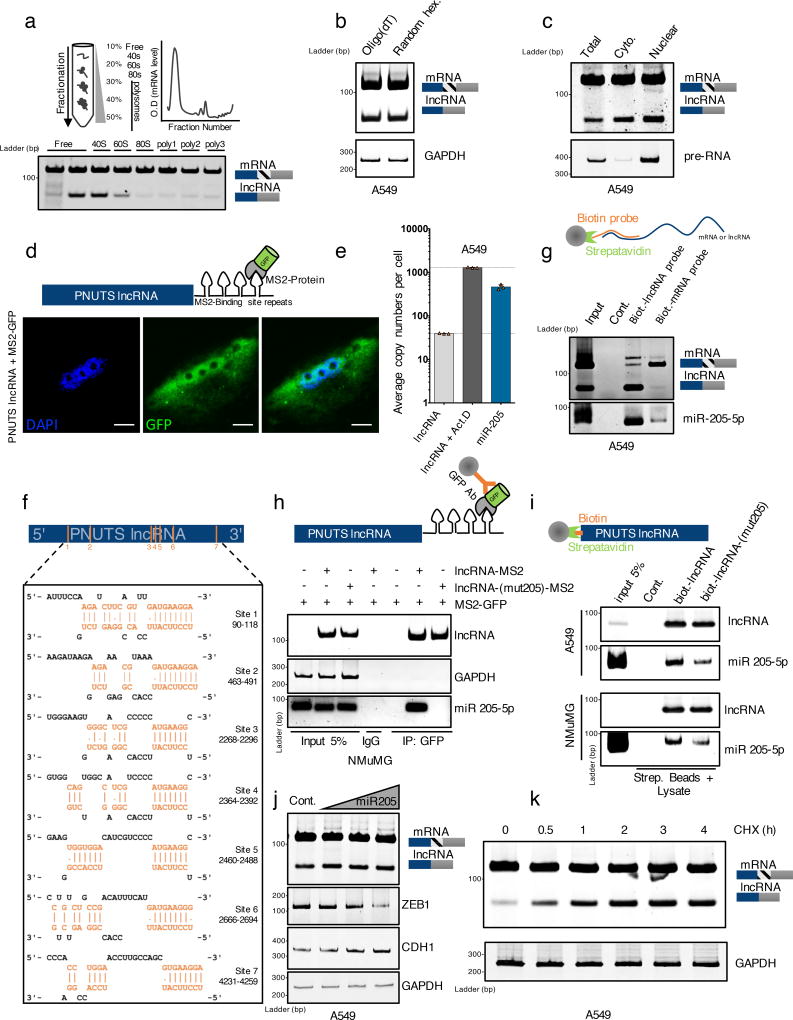
(a) Polysome fractionation experiment of A549 cells followed by RT-PCR analysis of PNUTS mRNA and lncRNA-PNUTS expression in each fraction.
(b) RT-PCR analysis of PNUTS mRNA and lncRNA-PNUTS expression after the use of oligo-(dT) or random hexanucleotides as primers for initial reverse transcription reaction.
(c) RT-PCR analysis of lncRNA-PNUTS expression in A549 cells. The total, cytoplasmic (Cyto.) and nuclear fractions are shown. PNUTS pre-RNA and PNUTS mRNA were used as endogenous controls to monitor the fractions purity.
(d) Confocal microscopy imaging of subcellular localization of lncRNA-PNUTS using co-transfection of a MS2-tagged-RNA construct of lncRNA-PNUTS and a fused MS2-GFP protein construct. Scale bar: 5µM.
(e) The exact copy numbers of lncRNA-PNUTS (basal levels or levels following activation by Actinomycin D treatment for 3h) and miR-205 were quantified with limiting-dilution qRT-PCR. Data are shown as mean ± s.d., n= 3 independent experiments per condition. Source data are available in Supplementary table 2.
(f) In silico prediction of MiR-205 binding sites located on lncRNA-PNUTS, obtained using the DIANA-microT web server.
(g) Selective pull-down of either endogenous lncRNA-PNUTS or PNUTS-mRNA isoforms by using antisense biotinylated probes followed by miRNA-specific RT-PCR analysis to detect endogenously associated miR-205 with lncRNA-PNUTS in A549 cells.
(h) MS2-RIP followed by miRNA-specific RT-PCR analysis to detect the association of miR-205 with lncRNA-PNUTS in NMuMG cells. LncRNA-PNUTS and GAPDH expression were used as internal controls.
(i) A549 and NMUMG cell lysates incubated with in vitro transcribed biotin-labeled lncRNA-PNUTS were subjected to pull-down followed by miRNA extraction and analysis by RT-PCR.
(j) A549 cells overexpressing lncRNA-PNUTS were transfected with an increasing concentration of a synthetic miR-205 mimic and the lncRNA expression was assessed by RT-PCR. ZEB-1 and CDH1 were used to monitor the efficiency of miR-205 overexpression on mesenchymal-epithelial transition (MET) process.
(k) Time course experiment by using RT-PCR analysis of lncRNA-PNUTS levels upon addition of 10µg.mL−1 cycloheximide in A549 cells.
GAPDH was used as a loading control.