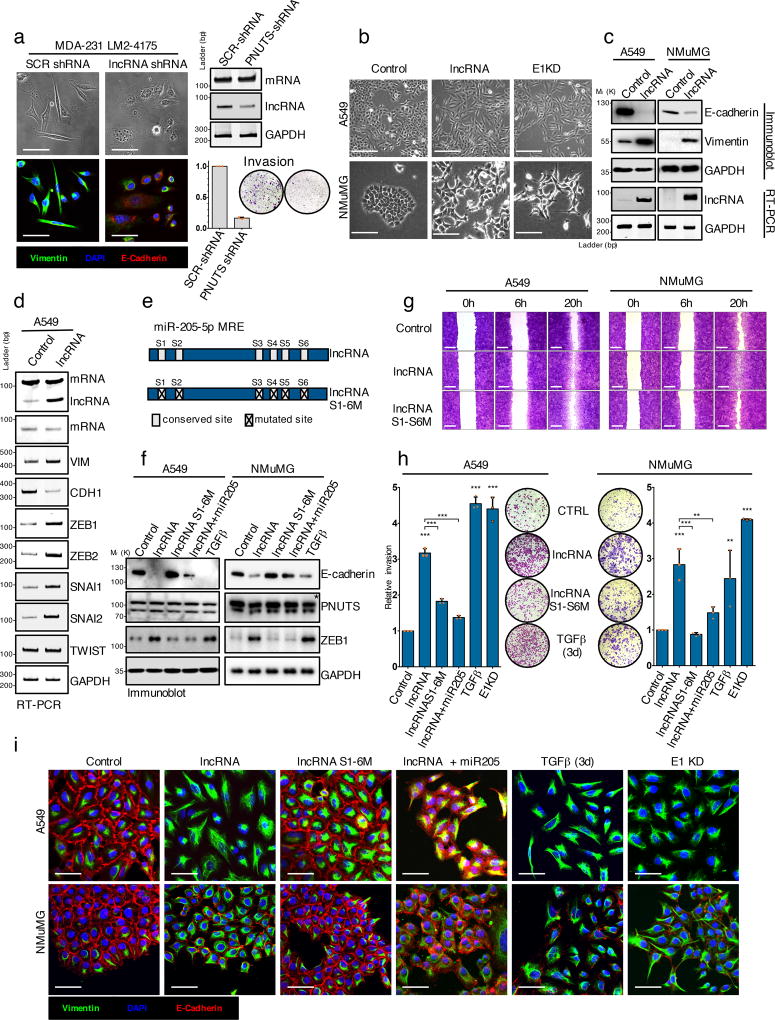
Figure 4. LncRNA-PNUTS regulates EMT and cell migration/invasion in vitro.
(a) MDA-231-LM2-4175 cells stably silenced for lncRNA-PNUTS were analyzed by immunofluorescence (left) using antibodies against vimentin (green), E-cadherin (red) and merged with DAPI (blue). Scale bar: 50µM. lncRNA-PNUTS silencing was monitored by RT-PCR (right, top). Invasive capacities of control (SCR-shRNA) or lncRNA-PNUTS silenced (PNUTS shRNA) cells were monitored in modified Boyden chamber assay (right, bottom). (Mean ± s.d., n= 3 independent experiments per condition). Source data are available in Supplementary table 2.
(b) A549 and NMuMG cells stably overexpressing lncRNA-PNUTS were analyzed using bright-field microscopy. hnRNP E1 knockdown (E1KD) cells were used as controls. Scale bar: 100µM.
(c) Western-blot (top) and RT-PCR (bottom) analysis of E-cadherin, vimentin and lncRNA-PNUTS in A549 and NMuMG cells overexpressing lncRNA-PNUTS.
(d) RT-PCR analysis of several EMT-related transcription factors in A549 cells stably overexpressing lncRNA-PNUTS.
(e) Schematic outlining the constructs used in this study for wild-type (lncRNA) or mutated (lncRNAS1-6M) lncRNA.
(f) Western-blot analysis of E-cadherin, PNUTS and ZEB1 protein expression in A549 and NMuMG cells overexpressing wild-type (lncRNA) or mutated (lncRNAS1-6M) constructs of lncRNA-PNUTS and treated or not with synthetic miR-205 mimic or TGFβ for 3 days. TGFβ was used as a positive control. * PNUTS protein band.
(g) Wound-healing migration assay of control (CTRL), lncRNA-PNUTS (lncRNA) or mutated lncRNA-PNUTS (lncRNAS1-6M) A549 and NMuMG cell models. Scale bar: 400µM
(h) Modified Boyden chamber invasion assay of wild-type (lncRNA) or mutated (lncRNAS1-6M) lncRNA-PNUTS overexpressing A549 and NMuMG cells pre-treated +/− synthetic miR-205 mimic or TGFβ for 3 days. hnRNP E1 knockdown (E1KD) and TGFβ treated cells were used as a positive control. (Mean ± s.d., n= 3 independent experiments per condition, ANOVA followed by post-hoc Tukey's multiple comparisons test, *p<0.05; **p<0.01; ***p<0.001, NS, not significant). Source data are available in Supplementary table 2.
(i) Confocal microscopy imaging of co-immunostaining of vimentin (green), E-cadherin (red) and merged with DAPI (blue) in A549 and NMuMG cells overexpressing wild-type (lncRNA) or mutated (lncRNAS1-6M) constructs of lncRNA-PNUTS and treated +/− synthetic miR-205 mimic or TGFβ for 3 days. Scale bar: 50µM.
For all western-blots and RT-PCRs GAPDH was used as a loading control. Unprocessed original scans of blots are shown in Supplementary Fig7