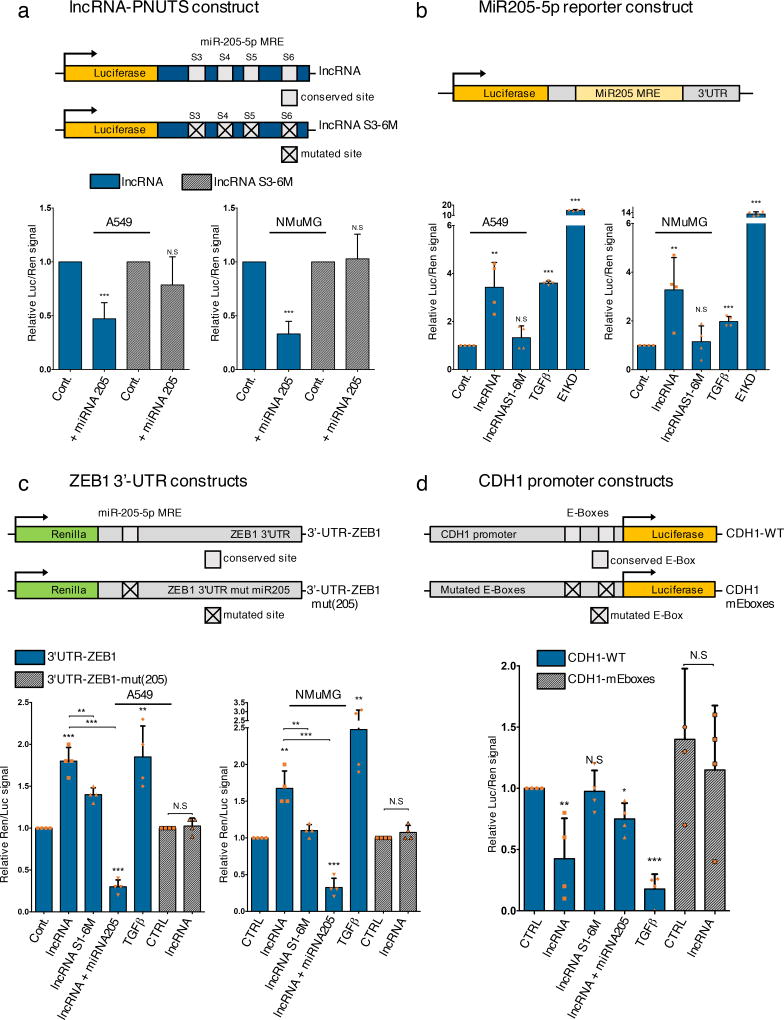
Figure 5. LncRNA-PNUTS controls the miR-205/ZEB/E-Cadherin axis.
(a) Dual luciferase reporter assays to test the interaction between miR-205 and lncRNA-PNUTS (S3 to S6 region) by using a synthetic miR-205 mimic (+ miRNA 205) co-transfected with wild-type (LUC-lncRNA) or mutated (LUC-lncRNA-S3-S6M) constructs of lncRNA-PNUTS cloned into the 3'-UTR of the luciferase reporter gene. For each condition, assays were normalized to Renilla reporter gene expression. (mean ± s.d., n= 7 independent experiments per condition, two-tailed Student t test, ***p<0.001, NS, not significant).
(b) Dual-Luciferase reporter assay of miR-205 bioavailability in A549 and NMuMG cells overexpressing wild-type (lncRNA) or mutated (lncRNAS1-6M) constructs of lncRNA-PNUTS. TGFβ treatment and hnRNP E1 knockdown (E1KD) were used as internal controls. For each condition, assays were normalized to Renilla reporter gene expression. (mean ± s.d., n= 4 independent experiments per condition, two-tailed Student t test, **p<0.01; ***p<0.001, NS, not significant).
(c) The wild-type (REN-3'-UTR-ZEB1) 3'-UTR of ZEB1 cloned into the 3'-UTR of the Renilla gene was transfected in A549 and NMuMG cells overexpressing wild-type (lncRNA) or mutated (lncRNAS1-6M) constructs of lncRNA-PNUTS and treated +/− synthetic miR-205 mimic or TGFβ for 3 days. Mutated construct (REN-3'-UTR-ZEB1-mut(205)) for the miR-205 binding site located in the 3'-UTR of ZEB1 was also used. TGFβ was used as a positive control. For each condition, assays were normalized to Luciferase reporter gene expression. (mean ± s.d., n= 4 independent experiments per condition, two-tailed Student t test, **p<0.01; ***p<0.001, NS, not significant).
(d) The wild-type (prom-CDH1-WT) proximal promoter of E-Cadherin driving the luciferase reporter gene expression was transfected in A549 cells overexpressing wild-type (lncRNA) or mutated (lncRNAS1-6M) constructs of lncRNA-PNUTS and treated or not with synthetic miR-205 mimics or TGFβ for 3 days. Mutated construct for both E2 Boxes 1 and 3 (Prom-CDH1-mEboxes) located on the promoter was also used. TGFβ was used as a positive control. For each condition, assays were normalized to Renilla reporter gene expression. (mean ± s.d., n= 4 independent experiments per condition, two-tailed Student t test, *p<0.05; **p<0.01; ***p<0.001, NS, not significant).