Abstract
Bacterial diversity underpins many ecosystem functions; however, the impact of within-species variation on the relationship between diversity and function remains unclear. Processes involving strain differentiation, such as niche radiation, are often overlooked in studies that focus on phylogenetic variation. This study used bacterial isolates assembled in two comparable microcosm experiments to test how species variation affected ecosystem function. We compared the relationship between diversity and activity (CO2 production) in increasingly diverse multispecies microcosms and with multiple ecotypes of a single species. The bacteria used were isolated from a low-diversity environment and are species of potential clinical significance such as Pseudomonas aeruginosa. All isolates were profiled for single carbon source utilisation. These data showed an increased breadth of resource use in the multiple ecotypes when compared to the mixed-species. The study observed significantly increasing respiration in more complex mixed-species assemblages, which was not observed when ecotypes of a single species were combined. We further demonstrate that the variation observed in the bacterial activity was due to the roles of each of the constituent isolates; between different species, the interactions between the isolates drove the variation in activity, whilst in single species, assemblage variation was due to which isolates were present. We conclude that both between- and within-species variations play different roles in community function, although through different mechanisms, and should be included in models of changing diversity and ecosystem functioning.
Electronic supplementary material
The online version of this article (doi:10.1007/s00248-017-0982-2) contains supplementary material, which is available to authorized users.
Keywords: Pseudomonas aeruginosa, Niche radiation, Biodiversity-ecosystem function, Intraspecific variation, Bacterial consortia, Carbon utilisation
Introduction
Whilst microbial communities are often highly phylogenetically and functionally diverse, much of the variation in these communities is masked at the sub-species level [1]. Intraspecific variation though is important and forms one mechanism by which single species can come to proliferate in environments, for example by radiating specialists into a broad range of niches thereby reducing competition and increasing complementary interactions [2–5]. Many current studies infer specific functions to individual species within communities as extrapolated from high throughput sequencing data, e.g. ([6]). Addressing the impact of intraspecific interaction and variation is, however, extremely challenging using such approaches. Manipulative studies, which give researchers the power to create experimental bacterial consortia, offer important alternative approaches through which ecosystem function can be related to changes in diversity [7]. Such biodiversity-ecosystem function (BEF) experiments have been reported in which bacterial species have been selected on morphological, phylogenetic or functional criteria [7–10]. Here, we selected bacteria based on data from both 16S rRNA gene sequence and single carbon source usage to set up two concurrent BEF experiments. The first BEF experiment analysed between-species interactions among phylogenetically diverse species, and the second studied within-species interactions between phylogenetically indistinguishable yet functionally diverse ecotypes [1] of the same species. This approach allowed us to conduct an investigation into how the “unseen” variation within a species can affect ecosystem functioning.
Bacteria were isolated from expectorated sputum sampled from individuals with cystic fibrosis (CF). CF respiratory infections are typically chronic by adulthood, with relatively low bacterial species diversity, high bacterial counts and varied habitats. In these conditions, BEF experiments are especially applicable [11–13]. This environment has also been observed to enable the adaptive radiation of the clinically important bacterial species Pseudomonas aeruginosa [4]. A total of 23 isolates were chosen of which 12 were identified as P. aeruginosa. The remaining 11 identified as a species from one of nine commonly isolated genera that naturally co-occur, representing the abundance of these genera observed in the sputum samples used (Table S1). Microcosms were assembled as described by the random partitions design [14]; within each set of mixed-species and mixed-ecotype combinations, a total of 28 microcosms were created containing either 1, 2, 3, 4, 6 or 12 isolates, with each isolate present once at each richness level. Two pools of isolates were used in this study, one using a mixture of 12 bacterial species and the other using 12 phylogenetically indistinguishable P. aeruginosa ecotypes (one P. aeruginosa isolate was used in both pools). These bacterial combinations were grown for 24 h at 37 °C statically in 30 mg mL−1 tryptone water. Respiration was measured using the MicroResp™ system and abundances of each isolate in the mixed-species microcosms were monitored through 16S rRNA gene terminal restriction fragment (T-RF) profiling from DNA extracted from the microcosms [15] (see Supplementary Methods).
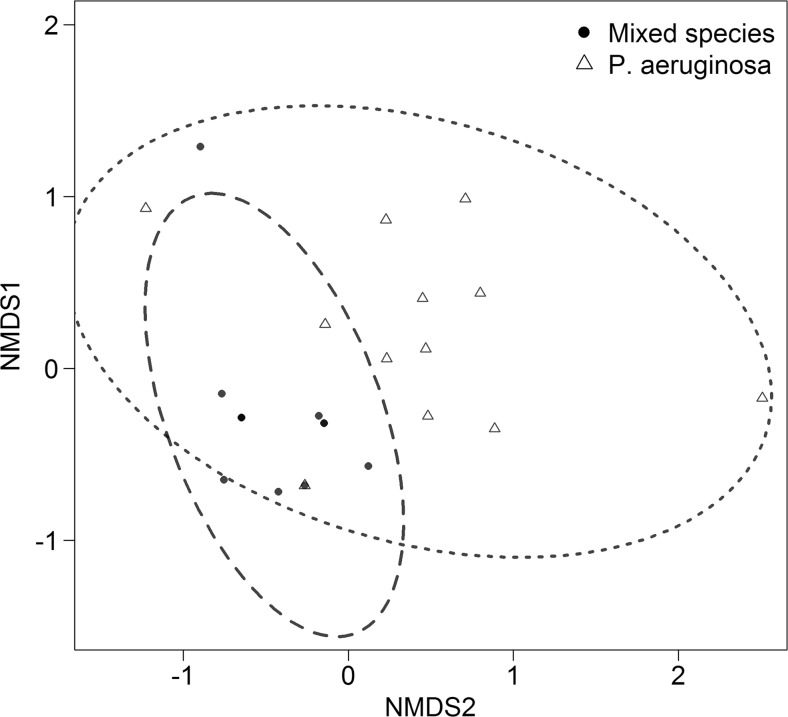
Fig. 2.
Ecotypes of a single species are more functionally dissimilar than phylogenetically diverse bacterial species. The ability of each isolate to utilise a range of carbon sources was used to determine that the different species (black circle) were more functionally similar than the P. aeruginosa ecotypes (white triangle) using Jaccard’s dissimilarity based on a binary presence/absence matrix. Dotted and dashed lines represent the 95% confidence interval around the mean distance within the group
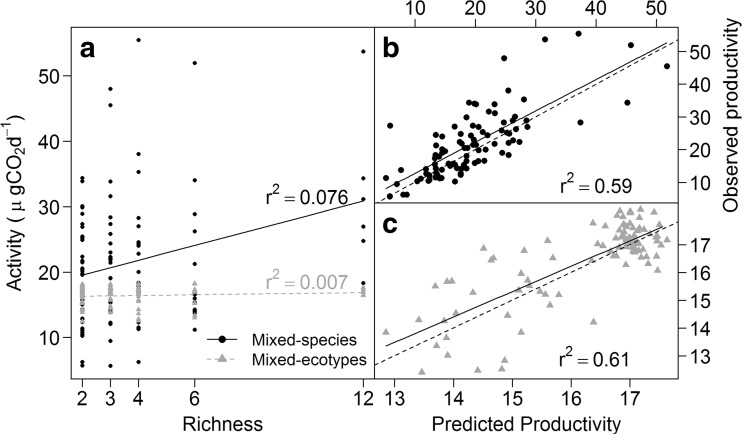
We observed (Fig. 1a) an increased respiration by 1.13 μg CO2 day−1 species−1 (F 1,10 = 8.77, P = 0.01) with increasing richness in the mixed-species BEF experiment. In contrast, in the mixed-ecotype experiment using Pseudomonads, there was no significant trend (0.04 μg CO2 day−1 ecotype−1, F 1,10 = 1.35, P = 0.27). Whilst no significant differences (t 71 = 1.40, P = 0.16) were observed between the mean monoculture activity values (±1 standard deviation throughout) of the mixed-species (17.53 ± 7.68 μg CO2 day−1) and P. aeruginosa ecotypes (16.24 ± 1.46 μg CO2 day−1), the mixed-species assemblages were consistently more active, and varied, than mixed-ecotype microcosms containing two or more isolates. This suggested that the positive trends observed in this mixed-species BEF experiment could be due to complementarity (niche differences or synergistic interactions) and as such not solely through selection effects [5].
Fig. 1.
Bacterial respiration is dependent on increasing phylogenetic richness rather than ecotypic richness. a A direct comparison of activity (μg CO2 day−1) of the microcosms with mixed-species ((black circle) black, solid line) and mixed-ecotypes only ((grey triangle) grey, dotted line). Community activity was shown to increase as more species were added to a microcosm; however, this effect was not observed as the number of P. aeruginosa ecotypes increased. This observed activity was shown to be predicted by the monocultures (solid black lines represent the trend of the data with the dotted black lines showing the 1:1 relationship for comparison) when considering both b mixed-species, in which the relative abundances were included and c mixed-ecotype microcosms, where activity was divided equally between the isolates
Further investigation of the data (Table 1) using a general linear model [14] found that the effect of bacterial interactions was both significant, greater significance was observed between mixed-species (F 4,10 = 15.91, P < 0.01) than mixed-ecotype assemblages (F 4,10 = 4.40, P = 0.02). We investigated this further by plotting the observed against the predicted activity for both BEF experiments (Fig. 1b, c) [10]. Here, the null model was equivalence between the observed and predicted values (1:1 relationship) with predicted values calculated using the total constituent monoculture activity divided by the number of species within an assemblage. We found that in the mixed-species assemblages (Fig. 1b), a significant difference between the prediction and the null model (β = 0.77, t 282 = 2.03, P = 0.043) was observed. This was eliminated when the relative abundances (T-RF profiles) were added into the predictions (β = 0.96, t 282 = 1.53, P = 0.127), thereby accounting for relative differences in species abundance [10]. Interactions between bacterial species are manifested by changes in their relative abundances [9], by both positive (e.g. cross-feeding [2]) and negative (e.g. competitive exclusion [16]) mechanisms. The prediction got closer to the null hypothesis with the inclusion of the interactions. This is not dependent solely on diversity but on the interplay between the species. By contrast, in P. aeruginosa ecotype assemblages, the null model prediction (Fig. 1c) was not significantly different from the null model (β = 0.91, t 282 = 0.07, P = 0.206), thereby suggesting that any differences in ecotype abundance did not impact upon overall activity.
Table 1.
Linear models estimating the effect of the additive model, composition and interactions on bacterial respiration. These biological factors were analysed with respect to the partitioned species pool and microcosm variance in the mixed-species and P. aeruginosa ecotype microcosms [14]. Residual degrees of freedom (Res.df) are those remaining after a variable has been added sequentially to the model
| Step | Variable | Res.df | F | P |
|---|---|---|---|---|
| Multispecies microcosms | ||||
| 1 | Respiration ~1 | 167 | – | – |
| 2 | 1+ additive model | 166 | 7.31 | 0.02 |
| 3 | 2+ composition | 154 | 1.1 | 0.38 |
| 4 | 3+ interactions | 150 | 15.91 | <0.01 |
| 5 | 4+ partitioned species pool | 138 | 3.14 | <0.01 |
| 6 | 5+ microcosm variation | 83 | 1.91 | <0.01 |
| P. aeruginosa microcosms | ||||
| 1 | Respiration ~1 | 167 | – | – |
| 2 | 1+ additive model | 166 | 0.14 | 0.71 |
| 3 | 2+ composition | 154 | 1.93 | 0.05 |
| 4 | 3+ interactions | 150 | 4.4 | 0.02 |
| 5 | 4+ partitioned species pool | 138 | 20.66 | <0.01 |
| 6 | 5+ microcosm variation | 83 | 1.19 | 0.26 |
Differences were found between the effects of composition (Table 1) in the ecotype and mixed-species experiments; composition did not significantly affect activity in mixed-species assemblages (F 11,55 = 1.10, P = 0.38); however, respiration was significantly affected by the different ecotype compositions (F 11,55 = 1.93, P = 0.05). To address why composition was important to the variation within the data, each isolate was tested using Biolog EcoPlates™ (Table S2) to assess their ability to utilise specific compounds as single sources of nutrients (analysed as a binary matrix). These results (Fig. 2) indicated that there were significant differences (ANOSIM R = 0.345, P = 0.01, 99 permutations) between the P. aeruginosa ecotypes (mean distance = 0.72 ± 0.17) and the mixed-species (0.44 ± 0.25). This suggested that the significant differences found for the P. aeruginosa ecotypes could be due to niche radiation. With the potential to radiate variation in niche usage within a given population, this is a plausible mechanism to explain the observed community dominance of P. aeruginosa during late-stage disease [4, 11]. It is interesting, however, that the combining ecotypes did not manage to effect activity as observed when mixing species. This was despite their apparent individual differences, as demonstrated by the carbon utilisation profiles, and the significant effect of composition on the activity values. The rationale for this cannot be elucidated in this study; however, we postulate that due to there being only one type of carbon source present in the media, the ecotypes may not have had the opportunity to interact and increase the overall activity. Further study is required to test our hypothesis.
In conclusion, we have shown that more mixed-species communities display more synergistic interactions, in terms of increased respiration, compared with assemblages of ecotypes from a single species. The differences between the mechanisms (species-species interactions vs. niche diversification) by which variation in the data arose illustrate different ways bacteria can affect overall community function. This study demonstrates the importance of including differing mechanisms of community dynamics when considering biodiversity-ecosystem function experiments; not only are interactions vital for the overyielding of mixed-species assemblages, but strain variation plays an interesting role within community ecology and should be accounted for in ecological analysis of BEF experiments and wider bacterial communities in nature.
Electronic Supplementary Material
(PDF 185 kb)
(DOCX 26 kb)
Acknowledgements
The authors would like to thank all of our colleagues who commented on this and previous versions of the manuscript. All sequences referred to in this study were deposited in Genbank under accession numbers KJ545900-KJ545922. The research reported here was undertaken under full ethical approval (NHS REC number: 08/H0502/126). This research and DWR were funded by a PhD studentship from SPARKS awarded to GJC, JPL and KDB.
Author Contributions
DWR, AKL and KDB conceived and designed the study. DWR performed the experiments and analysed the data with AKL. MPC collected the clinical samples. DWR and AKL wrote the manuscript which was completed with input from GJC, MPC, JPL and KDB.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s00248-017-0982-2) contains supplementary material, which is available to authorized users.
References
- 1.Cohan FM. What are bacterial species? Annu. Rev. Microbiol. 2002;56(1):457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence D, Fiegna F, Behrends V, et al. Species interactions alter evolutionary responses to a novel environment. PLoS Biol. 2012;10(5) doi: 10.1371/journal.pbio.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394(6688):69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 4.Smith EE, Buckley DG, Wu Z, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. PNAS. 2006;103(22):8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loreau M, Hector A. Partitioning selection and complementarity in biodiversity experiments. Nature. 2001;412(6842):72–76. doi: 10.1038/35083573. [DOI] [PubMed] [Google Scholar]
- 6.Muller EEL, Pinel N, Laczny CC, et al. Community-integrated omics links dominance of a microbial generalist to fine-tuned resource usage. Nat. Commun. 2014;5:5603. doi: 10.1038/ncomms6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell T, Gessner MO, Griffiths RI et al (2009) Microbial biodiversity and ecosystem functioning under controlled conditions and in the wild. In: Naeem S, Bunker DE, Hector A, Loreau M, Perrings C (eds) Biodiversity, ecosystem functioning, and human wellbeing: an ecological and economic perspective. Oxford University Press, pp. 121-133
- 8.Fiegna F, Moreno-Letelier A, Bell T, Barraclough TG. Evolution of species interactions determines microbial community productivity in new environments. ISME J. 2015;9(5):1235–1245. doi: 10.1038/ismej.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salles JF, Poly F, Schmid B, Roux XL. Community niche predicts the functioning of denitrifying bacterial assemblages. Ecology. 2009;90(12):3324–3332. doi: 10.1890/09-0188.1. [DOI] [PubMed] [Google Scholar]
- 10.Rivett DW, Scheuerl T, Culbert CT, et al. Resource-dependent attenuation of species interactions during bacterial succession. ISME J. 2016;10(9):2259–2268. doi: 10.1038/ismej.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goddard AF, Staudinger BJ, Dowd SE, et al. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. PNAS. 2012;109(34):13769–13774. doi: 10.1073/pnas.1107435109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stressmann FA, Rogers GB, Marsh P, et al. Does bacterial density in cystic fibrosis sputum increase prior to pulmonary exacerbation? J. Cyst. Fibros. 2011;10(5):357–365. doi: 10.1016/j.jcf.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Dickson RP, Erb-Downward JR, Freeman CM, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. 2015;12(6):821–830. doi: 10.1513/AnnalsATS.201501-029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell T, Lilley AK, Hector A, et al. A linear model method for biodiversity–ecosystem functioning experiments. Am. Nat. 2009;174(6):836–849. doi: 10.1086/647931. [DOI] [PubMed] [Google Scholar]
- 15.Rogers GB, Hart CA, Mason JR, et al. Bacterial diversity in cases of lung infection in cystic fibrosis patients: 16S ribosomal DNA (rDNA) length heterogeneity PCR and 16S rDNA terminal restriction fragment length polymorphism profiling. J. Clin. Microbiol. 2003;41(8):3548–3558. doi: 10.1128/JCM.41.8.3548-3558.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster KR, Bell T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr. Biol. 2012;22(19):1845–1850. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 185 kb)
(DOCX 26 kb)