Figure EV5. Establishment of a high‐resolution MNase‐ChIP‐seq protocol for Trypanosoma brucei .
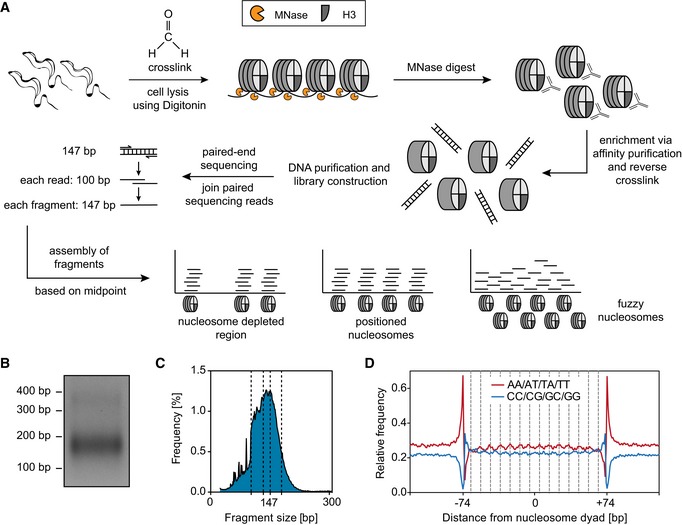
- Outline of MNase‐ChIP‐seq. T. brucei cells were formaldehyde‐cross‐linked and permeabilized, and chromatin was digested into mononucleosomes using MNase. Nucleosomes containing histone H3 were isolated via affinity purification using rabbit H3 antiserum. After reversing cross‐links, the nucleosomal DNA was purified and paired‐end‐sequenced using Illumina HiSeq 2500. The sequencing reads were joined to fragments and assembled according to their midpoints.
- 2% agarose gel with 100 ng of mononucleosomal DNA after an MNase digest.
- Fragment size distribution after sequencing and joining of paired sequencing reads. Dashed lines indicate the fragment sizes 100, 137, 147, and 157 bp.
- Relative frequencies of AA/AT/TA/TT and CC/CG/GC/GG dinucleotides throughout 147 bp of nucleosomal DNA for each bp relative to the nucleosome dyad. Dashed lines indicate distance of 10 bp from position −74 bp.
