Figure 1. Image‐based RNAi screening of RBR E3 ligases involved in bacterial Ub coat formation upon S. Typhimurium infection.
-
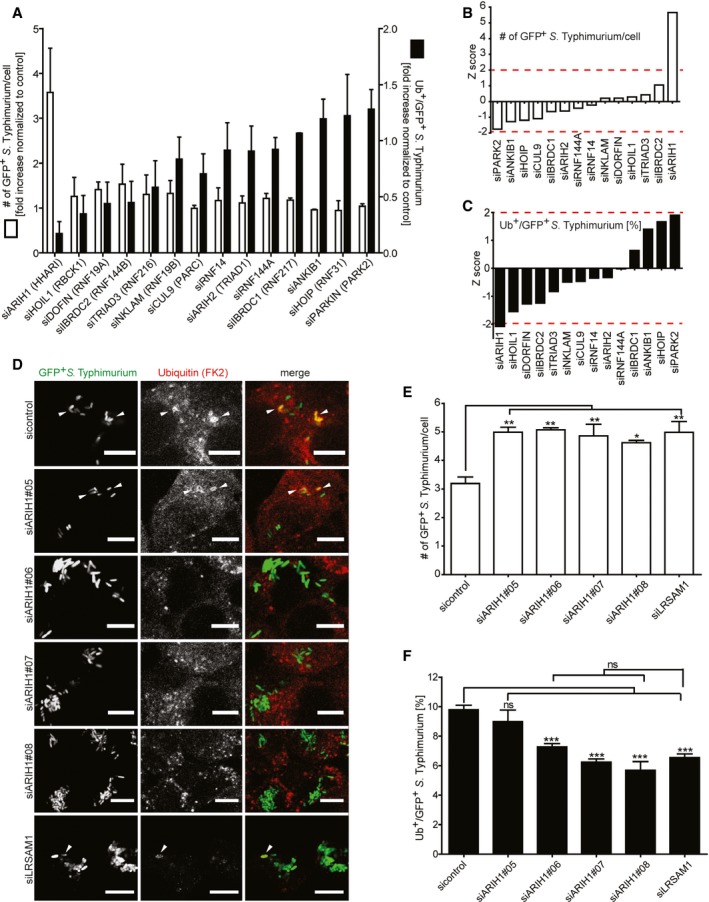
AHeLa cells reversely transfected for 72 h with non‐targeting control (sicontrol) or pooled siRNAs targeting all 14 known RBR Ub E3 ligases were infected with cytoGFP‐expressing ΔsifA S. Typhimurium for 2 h prior to fixation and immunolabeling with anti‐Ub antibody (FK2). Number of GFP‐positive (GFP+) and Ub‐ and GFP‐positive (Ub+/GFP+) bacteria was determined using automated quantification software and normalized to sicontrol counting on average 800 cells/sample (GFP‐positive S. Typhimurium/cell = 3.50 ± 0.13, ubiquitylated S. Typhimurium [%] = 8.32 ± 1.10). Data represent mean ± SD. n = 2 biological replicates.
-
B, Cz‐scores of GFP+ (B) and Ub+/GFP+ (C) bacteria from (A).
-
DHeLa cells were transfected with indicated single siRNAs, infected, fixed, and immunolabeled as in (A). Scale bar: 10 μm. Arrowheads indicate colocalization events.
-
E, FAutomated quantification of GFP+ (E) and Ub+/GFP+ (F) bacteria in on average 250 cells/sample. Data represent mean ± SD. Significance was determined using one‐way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001, ns = not significant. n = 3 biological replicates.
