Figure EV4. Gene phylogeny of histone H3 homologs.
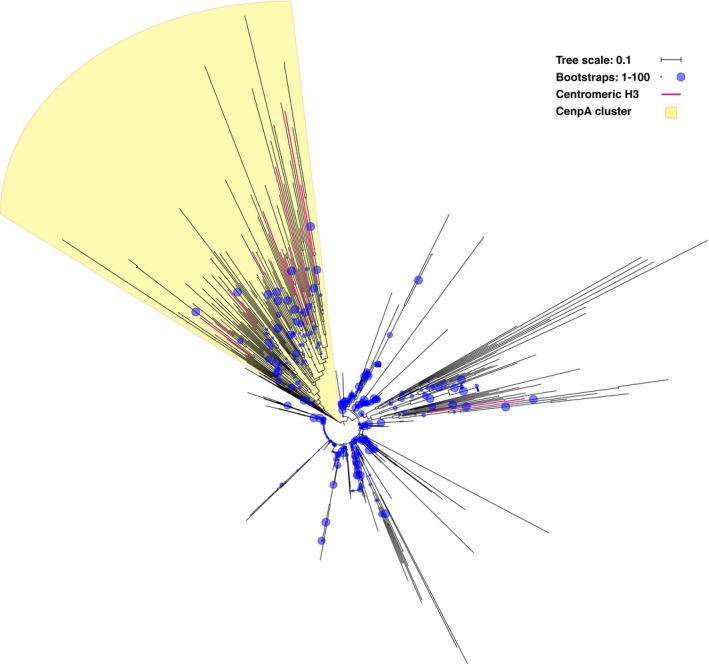
To find the putative orthologs of CenpA, we first aligned candidate orthologous sequences, which were experimentally identified centromeric H3 variants in divergent species (indicated with a pink branch in this phylogeny). From this alignment, we constructed a profile HMM and performed multiple HMM searches through our local proteome database. From these searches, we selected 831 sequences (belonging to the histone H3 family), aligned these and constructed the gene phylogeny, which is presented in this figure (see also Materials and Methods). We rooted the phylogeny on the cluster that contained all of these experimentally identified centromeric H3 variants and some additional sequences that, based on best blast hits, were also likely to be orthologous to CenpA. The cluster did not contain the candidate orthologs in Toxoplasma gondii 81. We do not know whether this is due to an error in the gene phylogeny, or to parallel invention of a centromeric H3 variants in this species, which would mean that it is not orthologous to CenpA. Nevertheless, we included these sequences in the orthologous group. The candidate centromeric H3 variants that are part of the CenpA cluster include sequences from all five eukaryotic supergroups: Homo sapiens 82, Saccharomyces cerevisiae 83, Drosophila melanogaster 84, Caenorhabditis elegans 85, Schizosaccharomyes pombe 86 (Opisthokonta), Dictyostelium discoideum 87 (Amoebozoa), Arabidopsis thaliana 88 (Archaeplastida), Tetrahymena thermophila 89, Plasmodium falciparum 90 (SAR), Giardia intestinalis 91 and Trichomonas vaginalis 92 (Excavata). The original gene tree in newick format is provided (Dataset EV3).
