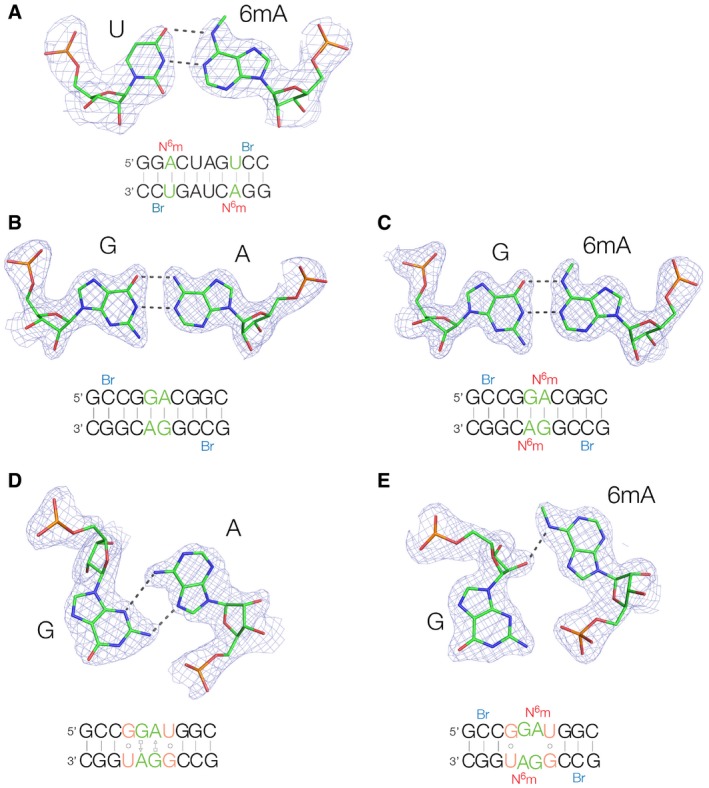
In each case, the relevant base pair is shown with its 2F
O‐F
C map contoured at 1.2σ. The sequences of each self‐complementary duplex are shown, with the relevant base pair highlighted in green, and N
6‐methyl modification in red. 5‐Bromocytosine nucleotides are shown blue; each structure was solved by SAD using the anomalous scatter from the two bromine atoms, except for 5LR3 that was determined by soaking with CuCl
2.
-
A
A duplex containing a N6‐methyladenine‐uracil base pair (PDB 5LR5). Crystals of space group P65 were obtained that diffracted to 2.27 Å. A standard cis Watson–Crick base pair is formed in this duplex, despite the presence of the N6‐methyl group.
-
B, C
Accommodation of N6‐methyladenine in a cis Watson–Crick G·A base pair. Two duplex species were constructed containing adjacent G·A base pairs flanked by G‐C base pairs, with (B, PDB 5LQO) and without (C, PDB 5LQT) N6‐methyladenine at 1.87 and 1.50‐Å resolution, respectively. In both structures, the G·A base pairs are cis Watson–Crick pairs connected by two hydrogen bonds. Thus, N6‐methylation of adenine does not lead to disruption of these Watson–Crick G·A pairs.
-
D, E
Disruption of a trans sugar‐Hoogsteen G·A base pair by N6‐methyladenine. Crystal structures of RNA duplexes in which guanine opposes adenine or N6‐methyladenine with flanking G·U base pairs. In the absence of the N6‐methyl group (D, PDB 5LR3 at 1.65‐Å resolution), a trans Hoogsteen‐sugar G·A base pair is formed. In marked contrast, the N6‐methyladenine does not form a base pair with the guanine (E, PDB 5LR4 at 1.72‐Å resolution), and there is no hydrogen bonding between the two nucleobases. Thus, N6‐methylation of adenine prevents the formation of the trans Hoogsteen‐sugar G·A base pair.