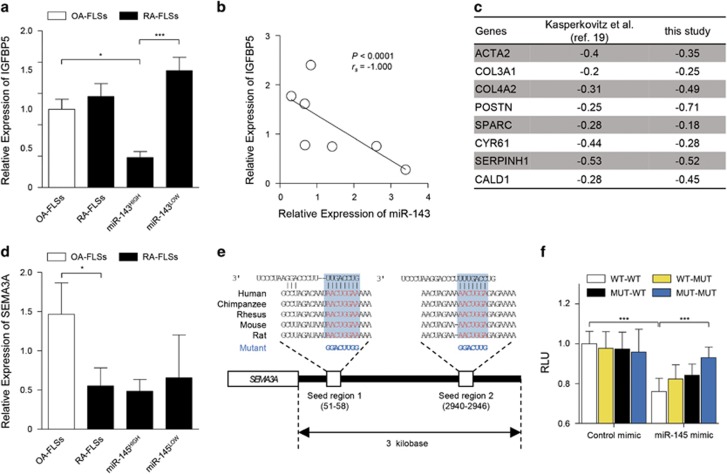
Figure 3.
Validation of IGFBP5 and SEMA3A as miR-143 and miR-145 targets, respectively. (a) Comparison of IGFBP5 mRNA expression between RA-FLSs (n=7) and OA-FLSs (n=10), which was determined by real-time PCR. GAPDH mRNA was used as an internal control. miR-143HIGH (n=3) and miR-143LOW (n=4) indicate RA-FLS subsets with higher and lower expression of miR-143, respectively. (b) Negative correlation between miR-143 and IGFBP5 expression in RA-FLSs (n=7). (c) Negative correlation of IGFBP5 gene expression levels with eight TFGβ-responsive genes in RA-FLSs (n=19) used in the previous study by Kasperkovitz et al.19 (GSE4061) and those (n=6) in this study (GSE22956 and GSE49604). ACTA2, Actin, Alpha 2, Smooth Muscle, Aorta; COL3A1, Collagen Type III Alpha 1 Chain; COL4A2, Collagen Type IV Alpha 2 Chain; POSTN, Periostin; SPARC, secreted protein acidic and rich in cysteine; CYR61, Cysteine Rich Angiogenic Inducer 61; SERPINH1, Serine (or Cysteine) Proteinase Inhibitor, Clade H (Heat Shock Protein 47), Member 1 (Collagen Binding Protein 1); CALD1, Caldesmon 1. Values are Pearson’s rho coefficients. (d) SEMA3A mRNA expression in RA-FLSs (n=7) versus OA-FLSs (n=10). miR-145HIGH (n=3) and miR-145LOW (n=4) indicate RA-FLS subsets with higher and lower expression of miR-145, respectively. (e) Sequence of miR-145 seed regions in the SEMA3A 3′-UTR. The mutation sequences are indicated in blue. (f) Relative normalized Renilla luciferase activity of reporter vector containing putative miR-145 binding sites (WT) or its mutated version (MUT). HEK293T cells were transfected with reporter vectors in the presence or absence of control mimic or synthetic miR-145 miRNA mimic. Single mutants of two putative miR-145-binding sites (seed regions) are indicated as WT-MUT or MUT-WT. A double mutant is denoted as MUT-MUT. RLU, relative light unit. The data show the mean±s.d. of three independent experiments. *P<0.05; ***P<0.001.