Figure 3.
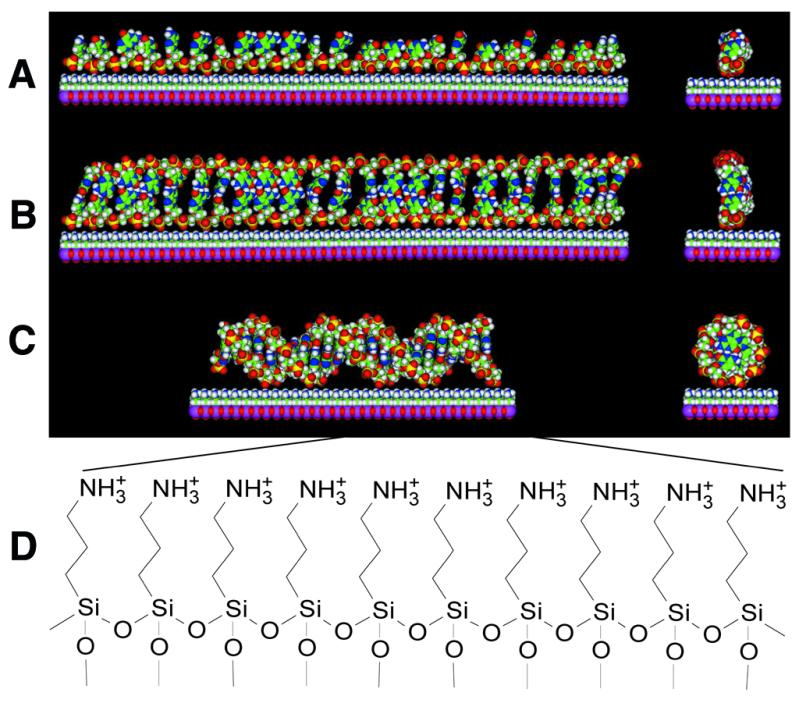
Models of duplex formation on a cationic surface. (A) A single-stranded 24mer oligonucleotide probe electrostatically attached to an aminosilanized surface. (B) A linear, non-helical DNA ribbon duplex on the surface, formed along its full length between an extended 24mer probe and its 24mer complementary target. (C) A 24 bp long B-form DNA double helix on the surface. (D) The chemical structure of the 3-aminopropyltrimethoxysilanized glass surface used in this study. The models in (A)–(C) were generated in the Molecular Builder module of InsightII and energy minimizations for ribbon duplex structure were done with the Amber force field of the Discover 3 package (MSI). Although it is quite possible that most common isomers of single-stranded (A) and double-stranded (B) DNA may have some curvature on a positively charged surface, only the linear isomers are shown for simplicity.
