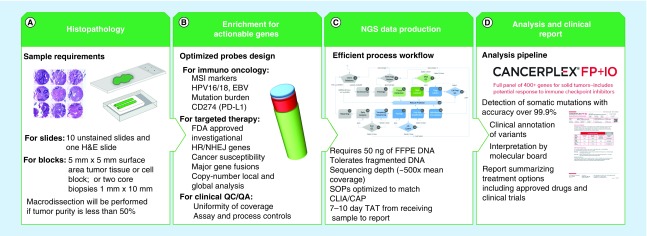
Figure 1. . Schematic presentation of CANCERPLEX® workflow.
The workflow is optimized to produce highly accurate and clinically relevant results from FFPE tumor tissue. (A) Slides are cut from tumor samples embedded in paraffin blocks, after which H&E staining is performed. Specimens that are composed of <50% tumor cells are dissected to extract regions with the greatest purity and viability of tumors cells. (B) Target enrichment probes have been custom designed to produce uniform coverage across all exons of the 435 targeted genes as well as regions of the genome that are likely to predict response to immuno-oncology drugs. (C) An overview of the process workflow is shown from sample acquisition to production of the patient's final report. (D) The raw sequencing data are uploaded to KEW proprietary informatics pipeline for identification of multiple classes of genetic variation, including SNVs, indels, CNVs, translocations, MSI and viral integration. The clinical significance of each genetic variant is determined by reviewing the scientific literature and cancer-related gene variant databases and FDA-approved and/or investigational therapies are reported.
CNV: Copy-number variant; FFPE: Formalin-fixed paraffin-embedded; H&E: Hematoxylin and eosin; indel: Insertion and deletion; MSI: Microsatellite instability; NGS: Next-generation sequencing; SNV: Single-nucleotide variant; TAT: Turnaround time.